Authors: Seth Billiau, Katherine Deng, Karissa Huang, and Sophia Li
This is a code appendix for our analysis of the Framingham Heart Study using GLMs and GAMS.
This is the code appendix for our EDA. The goals of this exercise were to discover missingness, visualize the distribution of the response and predictor variables, and assess the predictor variables for obvious signs of multicollinearity.
Read in the data and make note of missing values:
data_raw = read.csv("data/chd_risk.csv")
summary(data_raw)## age education cigsPerDay totChol
## Min. :32.00 College or Higher : 473 Min. : 0.000 Min. :107.0
## 1st Qu.:42.00 High School or GED:1253 1st Qu.: 0.000 1st Qu.:206.0
## Median :49.00 Some College : 687 Median : 0.000 Median :234.0
## Mean :49.58 Some High School :1720 Mean : 9.003 Mean :236.7
## 3rd Qu.:56.00 NA's : 105 3rd Qu.:20.000 3rd Qu.:263.0
## Max. :70.00 Max. :70.000 Max. :696.0
## NA's :29 NA's :50
## sysBP diaBP BMI heartRate
## Min. : 83.5 Min. : 48.00 Min. :15.54 Min. : 44.00
## 1st Qu.:117.0 1st Qu.: 75.00 1st Qu.:23.07 1st Qu.: 68.00
## Median :128.0 Median : 82.00 Median :25.40 Median : 75.00
## Mean :132.4 Mean : 82.89 Mean :25.80 Mean : 75.88
## 3rd Qu.:144.0 3rd Qu.: 89.88 3rd Qu.:28.04 3rd Qu.: 83.00
## Max. :295.0 Max. :142.50 Max. :56.80 Max. :143.00
## NA's :19 NA's :1
## glucose sex smoker OnBPMeds PrevStroke
## Min. : 40.00 female:2419 Nonsmoker:2144 No :4061 No :4213
## 1st Qu.: 71.00 male :1819 Smoker :2094 Yes : 124 Yes: 25
## Median : 78.00 NA's: 53
## Mean : 81.97
## 3rd Qu.: 87.00
## Max. :394.00
## NA's :388
## Hyp Diab CHD_Risk
## No :2922 No :4129 No :3594
## Yes:1316 Yes: 109 Yes: 644
##
##
##
##
##
length(data_raw$age)## [1] 4238
Count number of missing predictors in each variable:
# Generate the number of missing values for each predictor
apply(is.na(data_raw), 2, sum)## age education cigsPerDay totChol sysBP diaBP BMI
## 0 105 29 50 0 0 19
## heartRate glucose sex smoker OnBPMeds PrevStroke Hyp
## 1 388 0 0 53 0 0
## Diab CHD_Risk
## 0 0
missing_preds = c("education", "cigsPerDay", "totChol", "BMI",
"heartRate", "glucose", "OnBPMeds")# Labels for ticks
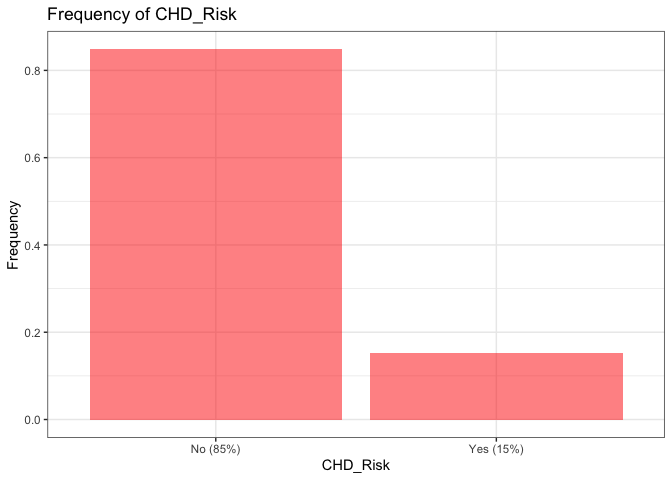
N_label = paste("No (", round(mean(data_raw$CHD_Risk == "No"), 2)*100, "%)", sep="")
Y_label = paste("Yes (", round(mean(data_raw$CHD_Risk == "Yes"), 2)*100, "%)", sep="")
# Plot bar chart
p <- ggplot(data=data_raw, aes(x=as.factor(CHD_Risk))) +
geom_histogram(stat="count",fill="red",
aes(y=..count../sum(..count..)),
alpha = 0.5) +
labs(title="Frequency of CHD_Risk") +
xlab("CHD_Risk") +
ylab("Frequency") +
scale_x_discrete(breaks=c("No","Yes"), labels=c(N_label,Y_label)) +
theme_bw()## Warning: Ignoring unknown parameters: binwidth, bins, pad
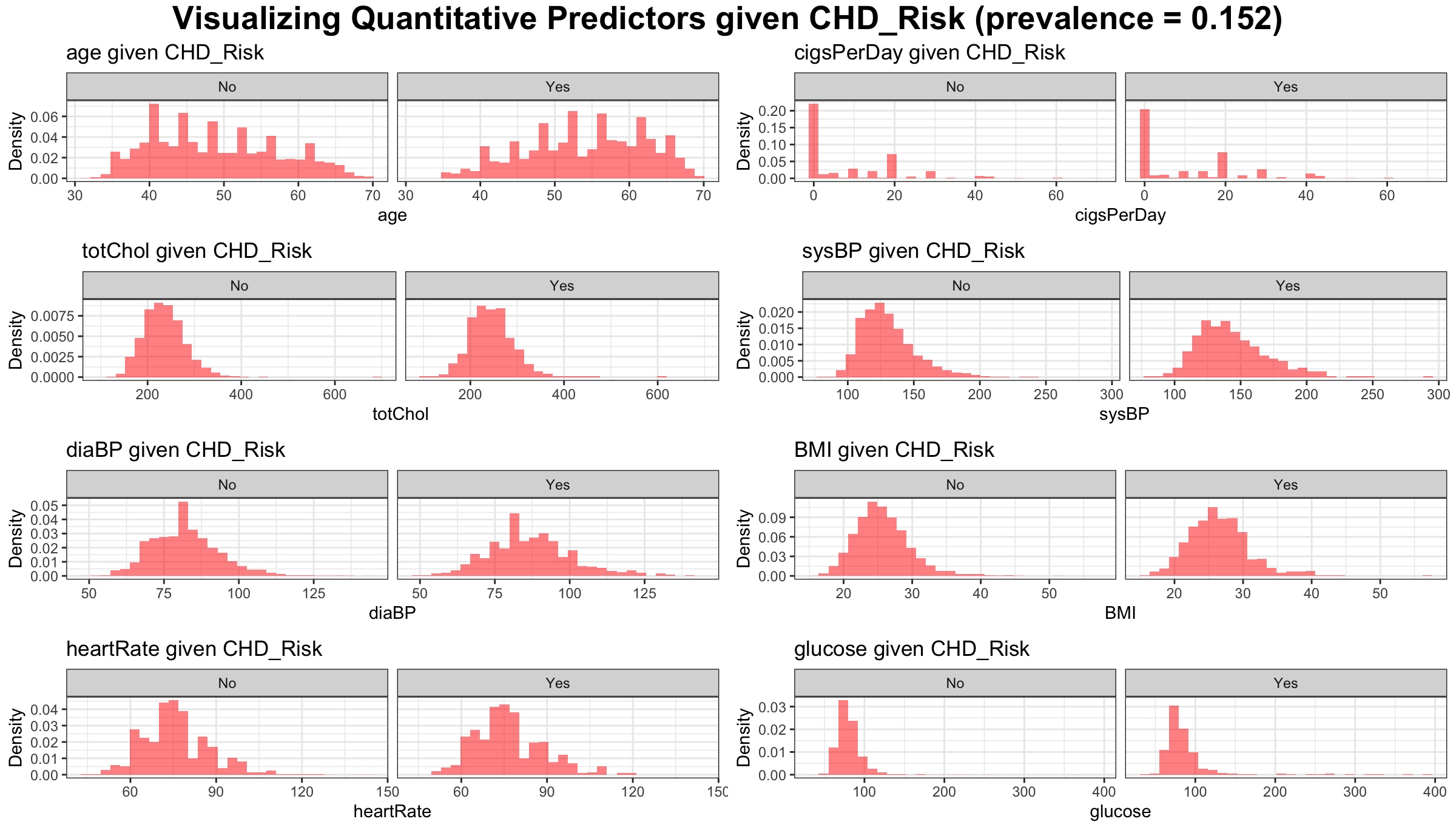
p Visualize distribution of quantitative predictors conditional on the CHD outcome:
quant_preds = c("age", "cigsPerDay", "totChol", "sysBP",
"diaBP", "BMI", "heartRate", "glucose")
make_cond_hist = function(varname) {
p1 = ggplot(data_raw, aes_string(x=varname)) +
geom_histogram(aes(y = ..density..),
fill = "red", alpha = 0.5) +
labs(title=paste(varname, "given CHD_Risk")) +
xlab(varname) +
ylab("Density") +
facet_grid(. ~ CHD_Risk) +
theme_bw()
return(p1)
}
graphs = lapply(quant_preds, make_cond_hist)
figure1 = ggarrange(graphs[[1]], graphs[[2]], graphs[[3]], graphs[[4]],
graphs[[5]], graphs[[6]], graphs[[7]], graphs[[8]],
ncol = 2, nrow = 4)
annotate_figure(figure1,
top = text_grob("Visualizing Quantitative Predictors given CHD_Risk (prevalence = 0.152)", face = "bold", size = 14)
)Visualizing the Qualitative predictors by showing their distributions conditional on the outcome:
# Address Categorical predictors
cat_preds = c("education", "sex", "smoker", "OnBPMeds",
"PrevStroke", "Hyp", "Diab")
get_cond_prob_table = function(TABLE) {
col1 = TABLE[,1] / sum(TABLE[,1])
col2 = TABLE[,2] / sum(TABLE[,2])
return(cbind(No=col1, Yes=col2))
}
tab_education = get_cond_prob_table(table(data_raw$education, data_raw$CHD_Risk))
tab_sex = get_cond_prob_table(table(data_raw$sex, data_raw$CHD_Risk))
tab_smoker = get_cond_prob_table(table(data_raw$smoker, data_raw$CHD_Risk))
tab_OnBPMeds = get_cond_prob_table(table(data_raw$OnBPMeds, data_raw$CHD_Risk))
tab_PrevStroke = get_cond_prob_table(table(data_raw$PrevStroke, data_raw$CHD_Risk))
tab_Hyp = get_cond_prob_table(table(data_raw$Hyp, data_raw$CHD_Risk))
tab_Diab = get_cond_prob_table(table(data_raw$Diab, data_raw$CHD_Risk))
tab_prob_Yes = rbind(tab_education, tab_sex, tab_smoker,
tab_OnBPMeds, tab_PrevStroke, tab_Hyp,
tab_Diab)
round(tab_prob_Yes,3)## No Yes
## College or Higher 0.115 0.111
## High School or GED 0.316 0.234
## Some College 0.171 0.140
## Some High School 0.399 0.514
## female 0.589 0.467
## male 0.411 0.533
## Nonsmoker 0.510 0.483
## Smoker 0.490 0.517
## No 0.977 0.935
## Yes 0.023 0.065
## No 0.996 0.983
## Yes 0.004 0.017
## No 0.724 0.495
## Yes 0.276 0.505
## No 0.981 0.938
## Yes 0.019 0.062
Check for collinearity with GVIF.
# Check for multicollinearity
mod.vif.lm <- lm(as.numeric(CHD_Risk) ~ ., data=data_raw)
vif(mod.vif.lm)## GVIF Df GVIF^(1/(2*Df))
## age 1.397737 1 1.182259
## education 1.124453 3 1.019742
## cigsPerDay 2.732416 1 1.653002
## totChol 1.116842 1 1.056808
## sysBP 3.767158 1 1.940917
## diaBP 3.000260 1 1.732126
## BMI 1.246685 1 1.116550
## heartRate 1.095015 1 1.046429
## glucose 1.638312 1 1.279966
## sex 1.223718 1 1.106218
## smoker 2.585357 1 1.607904
## OnBPMeds 1.111774 1 1.054407
## PrevStroke 1.017647 1 1.008785
## Hyp 2.051447 1 1.432287
## Diab 1.616622 1 1.271465
Because all values in the last column are less than 3.1623, there is not significant/strong evidence of multicollinearity.
This is the code appendix for our modeling section.
data = read.csv("data/chd_risk.csv")
# force smoker from categorical to numeric
data$smoker = unclass(data$smoker)
data_na = na.omit(data)
nrow(data_na)## [1] 3656
nrow(data)## [1] 4238
Fit a glm with individual predictors, smoker and cigPerDay are multiplied based on first principles.
glm_all = glm(CHD_Risk ~ age + education + totChol + sysBP + diaBP + BMI + heartRate + glucose + sex + smoker:cigsPerDay + OnBPMeds + PrevStroke + Hyp + Diab, family = binomial, data = data_na)
summary(glm_all)##
## Call:
## glm(formula = CHD_Risk ~ age + education + totChol + sysBP +
## diaBP + BMI + heartRate + glucose + sex + smoker:cigsPerDay +
## OnBPMeds + PrevStroke + Hyp + Diab, family = binomial, data = data_na)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -1.9363 -0.5940 -0.4232 -0.2837 2.8682
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -8.271975 0.699437 -11.827 < 2e-16 ***
## age 0.061962 0.006742 9.191 < 2e-16 ***
## educationHigh School or GED -0.131590 0.177573 -0.741 0.45866
## educationSome College -0.135339 0.197504 -0.685 0.49319
## educationSome High School 0.059549 0.164592 0.362 0.71750
## totChol 0.002362 0.001129 2.091 0.03650 *
## sysBP 0.015474 0.003810 4.061 4.88e-05 ***
## diaBP -0.004169 0.006441 -0.647 0.51745
## BMI 0.004476 0.012708 0.352 0.72471
## heartRate -0.002966 0.004211 -0.704 0.48120
## glucose 0.007211 0.002235 3.226 0.00126 **
## sexmale 0.534588 0.109948 4.862 1.16e-06 ***
## OnBPMedsYes 0.165180 0.234415 0.705 0.48103
## PrevStrokeYes 0.702450 0.491151 1.430 0.15266
## HypYes 0.233504 0.138183 1.690 0.09106 .
## DiabYes 0.025323 0.316114 0.080 0.93615
## smoker:cigsPerDay 0.010064 0.002117 4.753 2.00e-06 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 3120.5 on 3655 degrees of freedom
## Residual deviance: 2752.1 on 3639 degrees of freedom
## AIC: 2786.1
##
## Number of Fisher Scoring iterations: 5
Beginning analysis of deviance table. Decided not to do 1-term models with every predictor because that would take forever, so I used the Wald test predictors.
glm0 = glm(CHD_Risk ~ 1, family = binomial, data = data_na)
glm1 = glm(CHD_Risk ~ 1 + age, family = binomial, data = data_na)
glm2 = glm(CHD_Risk ~ 1 + sysBP, family = binomial, data = data_na)
glm3 = glm(CHD_Risk ~ 1 + sex, family = binomial, data = data_na)
glm4 = glm(CHD_Risk ~ 1 + smoker:cigsPerDay, family = binomial, data = data_na)
# age is the best
glm5 = glm(CHD_Risk ~ 1 + age + sysBP, family = binomial, data = data_na)
glm6 = glm(CHD_Risk ~ 1 + age + sex, family = binomial, data = data_na)
glm7 = glm(CHD_Risk ~ 1 + age + smoker:cigsPerDay, family = binomial, data = data_na)
# baseline age + sysBP
glm8 = glm(CHD_Risk ~ 1 + age + sysBP + sex, family = binomial, data = data_na)
glm9 = glm(CHD_Risk ~ 1 + age + sysBP + smoker:cigsPerDay, family = binomial, data = data_na)
# baseline age + sysBP + sex
glm10 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay, family = binomial, data = data_na)
# m working model
m = glm10
summary(glm10)##
## Call:
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay,
## family = binomial, data = data_na)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -1.5371 -0.5981 -0.4338 -0.2943 2.8106
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -8.251467 0.403019 -20.474 < 2e-16 ***
## age 0.068383 0.006346 10.776 < 2e-16 ***
## sysBP 0.018783 0.002127 8.829 < 2e-16 ***
## sexmale 0.537207 0.105146 5.109 3.24e-07 ***
## smoker:cigsPerDay 0.009354 0.002072 4.514 6.37e-06 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 3120.5 on 3655 degrees of freedom
## Residual deviance: 2785.6 on 3651 degrees of freedom
## AIC: 2795.6
##
## Number of Fisher Scoring iterations: 5
# anova(m, glm10, test = "Chisq")Continuing analysis of deviance - checking how each of the other 10 predictors adds to age, sex, sysBP, and smoker:cigsPerDay.
m = glm10
glm11 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + education, family = binomial, data = data_na)
glm12 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + totChol, family = binomial, data = data_na)
glm13 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + diaBP, family = binomial, data = data_na)
glm14 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + BMI, family = binomial, data = data_na)
glm15 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + heartRate, family = binomial, data = data_na)
glm16 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose, family = binomial, data = data_na)
glm17 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + OnBPMeds, family = binomial, data = data_na)
glm18 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + PrevStroke, family = binomial, data = data_na)
glm19 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + Hyp, family = binomial, data = data_na)
glm20 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + Diab, family = binomial, data = data_na)
# Should use a for loop here to examine deviance of each model
x = summary(glm20)
print(x$call)## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## Diab, family = binomial, data = data_na)
print(x$deviance)## [1] 2776.522
anova(glm16, m, test="Chisq")## Analysis of Deviance Table
##
## Model 1: CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose
## Model 2: CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay
## Resid. Df Resid. Dev Df Deviance Pr(>Chi)
## 1 3650 2766.5
## 2 3651 2785.6 -1 -19.102 1.239e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Baseline model: age, sysBP, sex, smoker:cigsPerDay, glucose.
Now, look for next predictor! Ignore diaBP and heartrate for now.
m = glm16
glm17 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + education, family = binomial, data = data_na)
glm18 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol, family = binomial, data = data_na)
glm19 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI, family = binomial, data = data_na)
glm20 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + OnBPMeds, family = binomial, data = data_na)
glm21 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + PrevStroke, family = binomial, data = data_na)
glm22 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + Hyp, family = binomial, data = data_na)
glm23 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + Diab, family = binomial, data = data_na)
pred6 <- list(glm17, glm18, glm19, glm20, glm21, glm22, glm23)
for (model_i in pred6){
print(model_i$call)
print(model_i$deviance)
}## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + education, family = binomial, data = data_na)
## [1] 2763.365
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol, family = binomial, data = data_na)
## [1] 2762.476
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + BMI, family = binomial, data = data_na)
## [1] 2766.02
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + OnBPMeds, family = binomial, data = data_na)
## [1] 2765.325
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + PrevStroke, family = binomial, data = data_na)
## [1] 2764.056
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + Hyp, family = binomial, data = data_na)
## [1] 2763.422
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + Diab, family = binomial, data = data_na)
## [1] 2766.465
anova(m, glm18, test = "Chisq")## Analysis of Deviance Table
##
## Model 1: CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose
## Model 2: CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose +
## totChol
## Resid. Df Resid. Dev Df Deviance Pr(>Chi)
## 1 3650 2766.5
## 2 3649 2762.5 1 4.0507 0.04415 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Working model is glm18: (age + sysBP + sex + smoker:cigsPerDay + glucose + totChol). Look for 7th predictors.
m = glm18
glm24 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + education, family = binomial, data = data_na)
glm25 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + OnBPMeds, family = binomial, data = data_na)
glm26 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + PrevStroke, family = binomial, data = data_na)
glm27 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + Hyp, family = binomial, data = data_na)
glm28 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + diaBP, family = binomial, data = data_na)
glm29 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + heartRate, family = binomial, data = data_na)
pred7 <- list(glm24, glm25, glm26, glm27, glm28, glm29)
for (model_i in pred7){
print(model_i$call)
print(model_i$deviance)
}## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + education, family = binomial, data = data_na)
## [1] 2758.95
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + OnBPMeds, family = binomial, data = data_na)
## [1] 2761.409
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + PrevStroke, family = binomial, data = data_na)
## [1] 2759.98
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + Hyp, family = binomial, data = data_na)
## [1] 2759.479
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + diaBP, family = binomial, data = data_na)
## [1] 2762.34
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + heartRate, family = binomial, data = data_na)
## [1] 2761.966
anova(m, glm24, test = "Chisq")## Analysis of Deviance Table
##
## Model 1: CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose +
## totChol
## Model 2: CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose +
## totChol + education
## Resid. Df Resid. Dev Df Deviance Pr(>Chi)
## 1 3649 2762.5
## 2 3646 2758.9 3 3.5264 0.3174
Interaction terms
int1 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + age:sysBP, family = binomial, data = data_na)
int2 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + age:sex, family = binomial, data = data_na)
int3 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + age:smoker:cigsPerDay, family = binomial, data = data_na)
int4 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + age:glucose, family = binomial, data = data_na)
int5 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + age:totChol, family = binomial, data = data_na)
int6 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + sysBP:sex, family = binomial, data = data_na)
int7 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + sysBP:smoker:cigsPerDay, family = binomial, data = data_na)
int8 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + sysBP:glucose, family = binomial, data = data_na)
int9 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + sysBP:totChol, family = binomial, data = data_na)
int10 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + sex:smoker:cigsPerDay, family = binomial, data = data_na)
int11 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + sex:glucose, family = binomial, data = data_na)
int12 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + sex:totChol, family = binomial, data = data_na)
int13 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + smoker:cigsPerDay:glucose, family = binomial, data = data_na)
int14 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + smoker:cigsPerDay:totChol, family = binomial, data = data_na)
int15 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol + glucose:totChol, family = binomial, data = data_na)
inter1 <- list(int1, int2, int3, int4, int5, int6, int7, int8, int9, int10, int11, int12, int13, int14, int15)
for (model_i in inter1){
print(model_i$call)
print(model_i$deviance)
}## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + age:sysBP, family = binomial, data = data_na)
## [1] 2761.898
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + age:sex, family = binomial, data = data_na)
## [1] 2762.233
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + age:smoker:cigsPerDay, family = binomial,
## data = data_na)
## [1] 2761.905
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + age:glucose, family = binomial, data = data_na)
## [1] 2762.468
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + age:totChol, family = binomial, data = data_na)
## [1] 2760.629
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + sysBP:sex, family = binomial, data = data_na)
## [1] 2760.044
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + sysBP:smoker:cigsPerDay, family = binomial,
## data = data_na)
## [1] 2762.39
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + sysBP:glucose, family = binomial, data = data_na)
## [1] 2762.476
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + sysBP:totChol, family = binomial, data = data_na)
## [1] 2762.268
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + sex:smoker:cigsPerDay, family = binomial,
## data = data_na)
## [1] 2761.693
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + sex:glucose, family = binomial, data = data_na)
## [1] 2762.258
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + sex:totChol, family = binomial, data = data_na)
## [1] 2759.904
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + smoker:cigsPerDay:glucose, family = binomial,
## data = data_na)
## [1] 2761.937
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + smoker:cigsPerDay:totChol, family = binomial,
## data = data_na)
## [1] 2762.429
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol + glucose:totChol, family = binomial, data = data_na)
## [1] 2759.187
anova(m, int15, test = "Chisq")## Analysis of Deviance Table
##
## Model 1: CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose +
## totChol
## Model 2: CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose +
## totChol + glucose:totChol
## Resid. Df Resid. Dev Df Deviance Pr(>Chi)
## 1 3649 2762.5
## 2 3648 2759.2 1 3.2892 0.06974 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
final.glm = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol, family = binomial, data = data_na)Examine model fit
summary(m)##
## Call:
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol, family = binomial, data = data_na)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -2.0074 -0.5966 -0.4315 -0.2864 2.8811
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -9.129843 0.475530 -19.199 < 2e-16 ***
## age 0.065896 0.006426 10.254 < 2e-16 ***
## sysBP 0.017534 0.002149 8.159 3.38e-16 ***
## sexmale 0.561446 0.106845 5.255 1.48e-07 ***
## glucose 0.007280 0.001677 4.342 1.41e-05 ***
## totChol 0.002272 0.001123 2.024 0.043 *
## smoker:cigsPerDay 0.009613 0.002088 4.604 4.14e-06 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 3120.5 on 3655 degrees of freedom
## Residual deviance: 2762.5 on 3649 degrees of freedom
## AIC: 2776.5
##
## Number of Fisher Scoring iterations: 5
3120.5 / 3655 # residual deviance / df## [1] 0.853762
# hosmer-lemeshow function (source: lowbwt-03.R)
hosmerlem = function (y, yhat, g = 10) {
cutyhat = cut(yhat, breaks = quantile(yhat, probs = seq(0, 1, 1/g)),
include.lowest = T)
obs = xtabs(cbind(1 - y, y) ~ cutyhat)
expect = xtabs(cbind(1 - yhat, yhat) ~ cutyhat)
chisq = sum((obs - expect)^2 / expect)
P = 1 - pchisq(chisq, g - 2)
c("X^2" = chisq, Df = g - 2, "P(>Chi)" = P)
}
# try different values of g to see if results are sensitive to choice
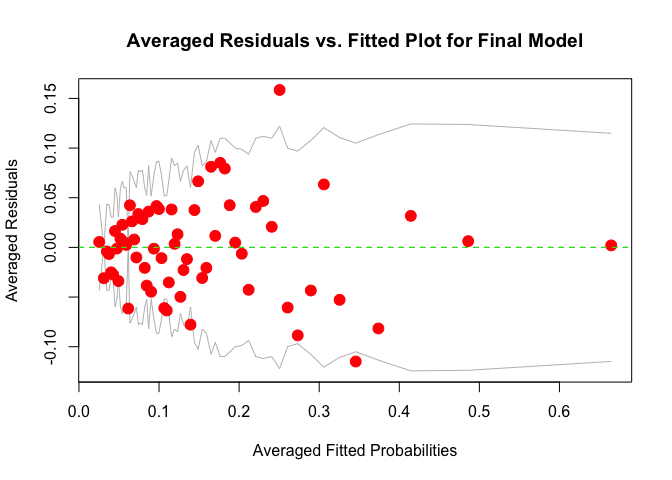
hosmerlem(m$y, fitted(m)) # g = 10## X^2 Df P(>Chi)
## 7.7444719 8.0000000 0.4588206
hosmerlem(m$y, fitted(m), g = 5) # g = 5## X^2 Df P(>Chi)
## 2.0660066 3.0000000 0.5588204
hosmerlem(m$y, fitted(m), g = 15) # g = 15## X^2 Df P(>Chi)
## 16.4053024 13.0000000 0.2279345
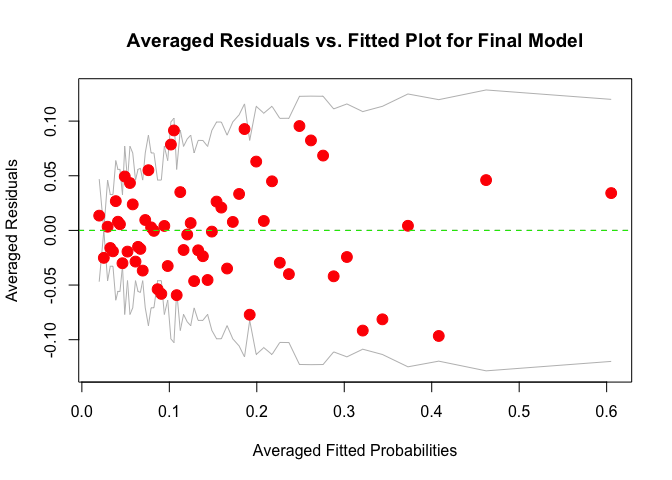
# plot averaged residuals vs. fitted probabilities
binnedplot(fitted(m), residuals(m, type = "response"),
xlab = "Averaged Fitted Probabilities",
ylab = "Averaged Residuals",
pch = 19, col.pts = "red", cex.pts = 1.5,
main = "Averaged Residuals vs. Fitted Plot for Final Model")
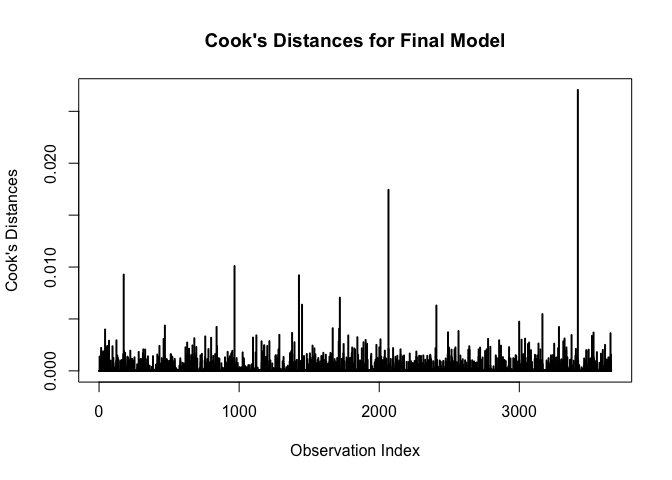
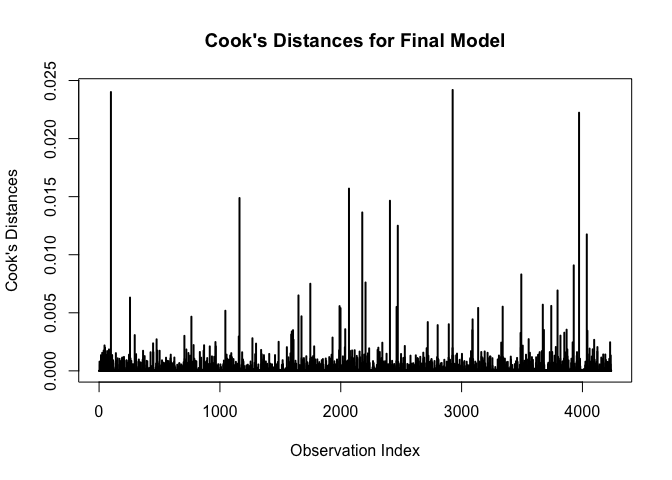
abline(h = 0, lty = 2, col = "green")# plot cook's distances
plot(cooks.distance(m), type = "h", lwd = 2,
xlab = "Observation Index",
ylab = "Cook's Distances",
main = "Cook's Distances for Final Model")
abline(h = 1, lty = 2, col = "red")Reprocess data:
# Begin building model
data = read.csv("data/chd_risk.csv")
# force smoker from categorical to numeric
data$smoker = unclass(data$smoker)
data_na = na.convert.mean(data)GLM with individual predictors, smoker and cigPerDay are combined based on first principles.
glm_all = glm(CHD_Risk ~ age + education + totChol + sysBP + diaBP + BMI + heartRate + glucose + sex + smoker:cigsPerDay + OnBPMeds + PrevStroke + Hyp + Diab + cigsPerDay.na + totChol.na + BMI.na + heartRate.na + glucose.na, family = binomial, data = data_na)
summary(glm_all)##
## Call:
## glm(formula = CHD_Risk ~ age + education + totChol + sysBP +
## diaBP + BMI + heartRate + glucose + sex + smoker:cigsPerDay +
## OnBPMeds + PrevStroke + Hyp + Diab + cigsPerDay.na + totChol.na +
## BMI.na + heartRate.na + glucose.na, family = binomial, data = data_na)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -1.9646 -0.5913 -0.4271 -0.2884 2.8544
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -7.879165 0.741040 -10.633 < 2e-16 ***
## age 0.060012 0.006292 9.538 < 2e-16 ***
## educationHigh School or GED -0.224931 0.165792 -1.357 0.17487
## educationNA -0.151727 0.316847 -0.479 0.63204
## educationSome College -0.116754 0.182979 -0.638 0.52343
## educationSome High School -0.023614 0.153580 -0.154 0.87780
## totChol 0.002054 0.001033 1.988 0.04681 *
## sysBP 0.014009 0.003567 3.927 8.59e-05 ***
## diaBP -0.002176 0.006012 -0.362 0.71734
## BMI 0.001837 0.011808 0.156 0.87634
## heartRate -0.001352 0.003902 -0.346 0.72909
## glucose 0.007098 0.002168 3.274 0.00106 **
## sexmale 0.498590 0.102658 4.857 1.19e-06 ***
## OnBPMedsNo -0.164830 0.362178 -0.455 0.64903
## OnBPMedsYes 0.085136 0.415618 0.205 0.83770
## PrevStrokeYes 0.829163 0.460535 1.800 0.07179 .
## HypYes 0.225356 0.129105 1.746 0.08089 .
## DiabYes 0.128827 0.297999 0.432 0.66552
## cigsPerDay.na -0.780859 0.747725 -1.044 0.29634
## totChol.na 0.222566 0.413593 0.538 0.59049
## BMI.na 2.131373 0.546400 3.901 9.59e-05 ***
## heartRate.na 12.377992 324.743752 0.038 0.96960
## glucose.na -0.098080 0.176282 -0.556 0.57795
## smoker:cigsPerDay 0.010847 0.001965 5.520 3.39e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 3611.5 on 4237 degrees of freedom
## Residual deviance: 3189.0 on 4214 degrees of freedom
## AIC: 3237
##
## Number of Fisher Scoring iterations: 11
Beginning analysis of deviance table. We decided not to do 1-term models with every predictor because that would take forever, so we used the Wald test predictors.
glm0 = glm(CHD_Risk ~ 1, family = binomial, data = data_na)
glm1 = glm(CHD_Risk ~ 1 + age, family = binomial, data = data_na)
glm2 = glm(CHD_Risk ~ 1 + sysBP, family = binomial, data = data_na)
glm3 = glm(CHD_Risk ~ 1 + sex, family = binomial, data = data_na)
glm4 = glm(CHD_Risk ~ 1 + smoker:cigsPerDay, family = binomial, data = data_na)
glm4b = glm(CHD_Risk ~ 1 + BMI.na, family = binomial, data = data_na)
pred1 <- list(glm0, glm1, glm2, glm3, glm4, glm4b)
for (model_i in pred1){
print(summary(model_i)$call)
print(summary(model_i)$deviance)
}## glm(formula = CHD_Risk ~ 1, family = binomial, data = data_na)
## [1] 3611.55
## glm(formula = CHD_Risk ~ 1 + age, family = binomial, data = data_na)
## [1] 3396.326
## glm(formula = CHD_Risk ~ 1 + sysBP, family = binomial, data = data_na)
## [1] 3432.642
## glm(formula = CHD_Risk ~ 1 + sex, family = binomial, data = data_na)
## [1] 3578.741
## glm(formula = CHD_Risk ~ 1 + smoker:cigsPerDay, family = binomial,
## data = data_na)
## [1] 3597.986
## glm(formula = CHD_Risk ~ 1 + BMI.na, family = binomial, data = data_na)
## [1] 3597.093
m = glm0
anova(m, glm1, test="Chisq")## Analysis of Deviance Table
##
## Model 1: CHD_Risk ~ 1
## Model 2: CHD_Risk ~ 1 + age
## Resid. Df Resid. Dev Df Deviance Pr(>Chi)
## 1 4237 3611.5
## 2 4236 3396.3 1 215.22 < 2.2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Age is the best predictor. Continue Analysis of deviance:
m = glm1
glm5 = glm(CHD_Risk ~ 1 + age + sysBP, family = binomial, data = data_na)
glm6 = glm(CHD_Risk ~ 1 + age + sex, family = binomial, data = data_na)
glm7 = glm(CHD_Risk ~ 1 + age + smoker:cigsPerDay, family = binomial, data = data_na)
glm7b = glm(CHD_Risk ~ 1 + age + BMI.na, family = binomial, data = data_na)
pred2 <- list(glm5, glm6, glm7, glm7b)
for (model_i in pred2){
print(summary(model_i)$call)
print(summary(model_i)$deviance)
}## glm(formula = CHD_Risk ~ 1 + age + sysBP, family = binomial,
## data = data_na)
## [1] 3325.673
## glm(formula = CHD_Risk ~ 1 + age + sex, family = binomial, data = data_na)
## [1] 3356.181
## glm(formula = CHD_Risk ~ 1 + age + smoker:cigsPerDay, family = binomial,
## data = data_na)
## [1] 3347.071
## glm(formula = CHD_Risk ~ 1 + age + BMI.na, family = binomial,
## data = data_na)
## [1] 3383.409
anova(m, glm5, test="Chisq")## Analysis of Deviance Table
##
## Model 1: CHD_Risk ~ 1 + age
## Model 2: CHD_Risk ~ 1 + age + sysBP
## Resid. Df Resid. Dev Df Deviance Pr(>Chi)
## 1 4236 3396.3
## 2 4235 3325.7 1 70.653 < 2.2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
sysBP is the best predictor. Continue Analysis of deviance:
m = glm5
# baseline age + sysBP
glm8 = glm(CHD_Risk ~ 1 + age + sysBP + sex, family = binomial, data = data_na)
glm9 = glm(CHD_Risk ~ 1 + age + sysBP + smoker:cigsPerDay, family = binomial, data = data_na)
glm9b = glm(CHD_Risk ~ 1 + age + sysBP + BMI.na, family = binomial, data = data_na)
pred3 <- list(glm8, glm9, glm9b)
for (model_i in pred3){
print(summary(model_i)$call)
print(summary(model_i)$deviance)
}## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex, family = binomial,
## data = data_na)
## [1] 3273.349
## glm(formula = CHD_Risk ~ 1 + age + sysBP + smoker:cigsPerDay,
## family = binomial, data = data_na)
## [1] 3272.317
## glm(formula = CHD_Risk ~ 1 + age + sysBP + BMI.na, family = binomial,
## data = data_na)
## [1] 3313.134
anova(m, glm9, test="Chisq")## Analysis of Deviance Table
##
## Model 1: CHD_Risk ~ 1 + age + sysBP
## Model 2: CHD_Risk ~ 1 + age + sysBP + smoker:cigsPerDay
## Resid. Df Resid. Dev Df Deviance Pr(>Chi)
## 1 4235 3325.7
## 2 4234 3272.3 1 53.356 2.782e-13 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
smoker:cigsPerDay is the best predictor. Continue Analysis of deviance:
m = glm9
# baseline age + sysBP + smoker:cigsPerDay
glm10 = glm(CHD_Risk ~ 1 + age + sysBP + smoker:cigsPerDay + sex, family = binomial, data = data_na)
glm10b = glm(CHD_Risk ~ 1 + age + sysBP + smoker:cigsPerDay + BMI.na, family = binomial, data = data_na)
for (i in list(glm10, glm10b)){
print(summary(i)$call)
print(summary(i)$deviance)
}## glm(formula = CHD_Risk ~ 1 + age + sysBP + smoker:cigsPerDay +
## sex, family = binomial, data = data_na)
## [1] 3245.906
## glm(formula = CHD_Risk ~ 1 + age + sysBP + smoker:cigsPerDay +
## BMI.na, family = binomial, data = data_na)
## [1] 3258.933
Continuing analysis of deviance - checking how each of the other 10 predictors adds to age, sex, sysBP, and smoker:cigsPerDay:
m = glm10
glm11 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + education, family = binomial, data = data_na)
glm12 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + totChol, family = binomial, data = data_na)
glm13 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + diaBP, family = binomial, data = data_na)
glm14 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + BMI, family = binomial, data = data_na)
glm15 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + heartRate, family = binomial, data = data_na)
glm16 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose, family = binomial, data = data_na)
glm17 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + OnBPMeds, family = binomial, data = data_na)
glm18 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + PrevStroke, family = binomial, data = data_na)
glm19 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + Hyp, family = binomial, data = data_na)
glm20 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + Diab, family = binomial, data = data_na)
glm20b = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + BMI.na, family = binomial, data = data_na)
pred5 <- list(glm11, glm12, glm13, glm14, glm15, glm16, glm17, glm18, glm19, glm20, glm20b)
for (model_i in pred5){
print(summary(model_i)$call)
print(summary(model_i)$deviance)
}
anova(glm16, m, test="Chisq")Baseline model: age, sysBP, sex, smoker:cigsPerDay, glucose.
Look for next predictor! Ignore diaBP and heartrate for now.
m = glm16
glm17 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + education, family = binomial, data = data_na)
glm18 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol, family = binomial, data = data_na)
glm19 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI, family = binomial, data = data_na)
glm20 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + OnBPMeds, family = binomial, data = data_na)
glm21 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + PrevStroke, family = binomial, data = data_na)
glm22 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + Hyp, family = binomial, data = data_na)
glm23 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + Diab, family = binomial, data = data_na)
glm23b = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na, family = binomial, data = data_na)
pred6 <- list(glm17, glm18, glm19, glm20, glm21, glm22, glm23, glm23b)
for (model_i in pred6){
print(model_i$call)
print(model_i$deviance)
}
anova(m, glm23b, test = "Chisq")Working model is glm23b: (age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na)
Look for 7th predictors:
m = glm23b
glm24 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + education, family = binomial, data = data_na)
glm25 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + OnBPMeds, family = binomial, data = data_na)
glm26 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke, family = binomial, data = data_na)
glm27 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + Hyp, family = binomial, data = data_na)
glm28 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + diaBP, family = binomial, data = data_na)
glm29 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + heartRate, family = binomial, data = data_na)
glm30 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + totChol, family = binomial, data = data_na)
pred7 <- list(glm24, glm25, glm26, glm27, glm28, glm29, glm30)
for (model_i in pred7){
print(model_i$call)
print(model_i$deviance)
}
anova(m, glm26, test = "Chisq")Working model is glm26: (age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke)
Look for 8th predictors:
m = glm26
glm31 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + education, family = binomial, data = data_na)
glm32 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + OnBPMeds, family = binomial, data = data_na)
glm33 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + Hyp, family = binomial, data = data_na)
glm34 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + Diab, family = binomial, data = data_na)
glm35 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + BMI.na, family = binomial, data = data_na)
glm36 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + totChol, family = binomial, data = data_na)
pred8 <- list(glm31, glm32, glm33, glm34, glm35, glm36)
for (model_i in pred8){
print(model_i$call)
print(model_i$deviance)
}
anova(m, glm31, test="Chisq")Working model is glm26: (age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke)
Look for two-way Interaction Terms:
# Interaction terms
int1 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + age:sysBP, family = binomial, data = data_na)
int2 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + age:sex, family = binomial, data = data_na)
int3 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + age:smoker:cigsPerDay, family = binomial, data = data_na)
int4 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + age:glucose, family = binomial, data = data_na)
int5 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + age:BMI.na, family = binomial, data = data_na)
int6 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + age:PrevStroke, family = binomial, data = data_na)
int7 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sysBP:sex, family = binomial, data = data_na)
int8 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sysBP:smoker:cigsPerDay, family = binomial, data = data_na)
int9 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sysBP:glucose, family = binomial, data = data_na)
int10 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sysBP:BMI.na, family = binomial, data = data_na)
int11 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sysBP:PrevStroke, family = binomial, data = data_na)
int12 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sex:smoker:cigsPerDay, family = binomial, data = data_na)
int13 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sex:glucose, family = binomial, data = data_na)
int14 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sex:BMI.na, family = binomial, data = data_na)
int15 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sex:PrevStroke, family = binomial, data = data_na)
int16 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + smoker:cigsPerDay:glucose, family = binomial, data = data_na)
int17 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + smoker:cigsPerDay:BMI.na, family = binomial, data = data_na)
int18 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + smoker:cigsPerDay:PrevStroke, family = binomial, data = data_na)
int19 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + glucose:BMI.na, family = binomial, data = data_na)
int20 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + glucose:PrevStroke, family = binomial, data = data_na)
int21 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + BMI.na:PrevStroke, family = binomial, data = data_na)
inter1 <- list(int1, int2, int3, int4, int5, int6, int7, int8, int9, int10, int11, int12, int13, int14, int15, int16, int17, int18, int19, int20, int21)
for (model_i in inter1){
print(model_i$call)
print(model_i$deviance)
}
anova(m, int7, test = "Chisq")Working model is int7: (age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sysBP:sex).
Look for two-way Interaction Terms:
m = int7
# Interaction terms
int22 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + BMI.na:PrevStroke + sex:sysBP, family = binomial, data = data_na)
int23 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + age:sex + sex:sysBP, family = binomial, data = data_na)
int24 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + age:smoker:cigsPerDay + sex:sysBP, family = binomial, data = data_na)
int25 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + age:glucose + sex:sysBP, family = binomial, data = data_na)
int26 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + age:BMI.na + sex:sysBP, family = binomial, data = data_na)
int27 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + age:PrevStroke + sex:sysBP, family = binomial, data = data_na)
int28 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sysBP:sex + sex:sysBP, family = binomial, data = data_na)
int29 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sysBP:smoker:cigsPerDay + sex:sysBP, family = binomial, data = data_na)
int30 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sysBP:glucose + sex:sysBP, family = binomial, data = data_na)
int31 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sysBP:BMI.na + sex:sysBP, family = binomial, data = data_na)
int32 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sysBP:PrevStroke + sex:sysBP, family = binomial, data = data_na)
int33 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sex:smoker:cigsPerDay + sex:sysBP, family = binomial, data = data_na)
int34 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sex:glucose + sex:sysBP, family = binomial, data = data_na)
int35 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sex:BMI.na + sex:sysBP, family = binomial, data = data_na)
int36 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + sex:PrevStroke + sex:sysBP, family = binomial, data = data_na)
int37 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + smoker:cigsPerDay:glucose + sex:sysBP, family = binomial, data = data_na)
int38 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + smoker:cigsPerDay:BMI.na + sex:sysBP, family = binomial, data = data_na)
int39 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + smoker:cigsPerDay:PrevStroke + sex:sysBP, family = binomial, data = data_na)
int40 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + glucose:BMI.na + sex:sysBP, family = binomial, data = data_na)
int41 = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + BMI.na + PrevStroke + glucose:PrevStroke + sex:sysBP, family = binomial, data = data_na)
inter1 <- list(m, int22, int23, int24, int25, int26, int27, int28, int29, int30, int31, int32, int33, int34, int35, int36, int37, int38, int39, int40, int41)
for (model_i in inter1){
print(model_i$call)
print(model_i$deviance)
}
anova(m, int35, test = "Chisq")End Analysis of Deviance test.
Examine model fit.
summary(m)##
## Call:
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + BMI.na + PrevStroke + sysBP:sex, family = binomial,
## data = data_na)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -2.1558 -0.5867 -0.4307 -0.3041 2.7586
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -8.103274 0.435259 -18.617 < 2e-16 ***
## age 0.065520 0.005970 10.975 < 2e-16 ***
## sysBP 0.014012 0.002550 5.495 3.91e-08 ***
## sexmale -0.597942 0.555410 -1.077 0.28167
## glucose 0.007771 0.001638 4.744 2.10e-06 ***
## BMI.na 2.087449 0.542855 3.845 0.00012 ***
## PrevStrokeYes 0.932917 0.452335 2.062 0.03917 *
## smoker:cigsPerDay 0.010819 0.001940 5.578 2.44e-08 ***
## sysBP:sexmale 0.007883 0.003926 2.008 0.04464 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 3611.5 on 4237 degrees of freedom
## Residual deviance: 3201.2 on 4229 degrees of freedom
## AIC: 3219.2
##
## Number of Fisher Scoring iterations: 5
3120.5 / 3655 # residual deviance / df## [1] 0.853762
# hosmer-lemeshow function (source: lowbwt-03.R)
hosmerlem = function (y, yhat, g = 10) {
cutyhat = cut(yhat, breaks = quantile(yhat, probs = seq(0, 1, 1/g)),
include.lowest = T)
obs = xtabs(cbind(1 - y, y) ~ cutyhat)
expect = xtabs(cbind(1 - yhat, yhat) ~ cutyhat)
chisq = sum((obs - expect)^2 / expect)
P = 1 - pchisq(chisq, g - 2)
c("X^2" = chisq, Df = g - 2, "P(>Chi)" = P)
}
# try different values of g to see if results are sensitive to choice
hosmerlem(m$y, fitted(m)) # g = 10## X^2 Df P(>Chi)
## 11.2533209 8.0000000 0.1877547
hosmerlem(m$y, fitted(m), g = 5) # g = 5## X^2 Df P(>Chi)
## 7.04608104 3.00000000 0.07044343
hosmerlem(m$y, fitted(m), g = 15) # g = 15## X^2 Df P(>Chi)
## 19.16186 13.00000 0.11817
Plot averaged residuals vs. fitted probabilities:
binnedplot(fitted(m), residuals(m, type = "response"),
xlab = "Averaged Fitted Probabilities",
ylab = "Averaged Residuals",
pch = 19, col.pts = "red", cex.pts = 1.5,
main = "Averaged Residuals vs. Fitted Plot for Final Model")
abline(h = 0, lty = 2, col = "green")Plot Cook’s distances:
plot(cooks.distance(m), type = "h", lwd = 2,
xlab = "Observation Index",
ylab = "Cook's Distances",
main = "Cook's Distances for Final Model")
abline(h = 1, lty = 2, col = "red")Initialize a GAM with all of our models.
gam.init <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp+Diab, family = binomial, data = data_na)
gam.age1 <- gam(CHD_Risk~education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp+Diab, family = binomial, data = data_na)
gam.education1 <- gam(CHD_Risk~s(age)+s(cigsPerDay,smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp+Diab, family = binomial, data = data_na)
gam.totChol1 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp+Diab, family = binomial, data = data_na)
gam.sysBP1 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay,smoker)+s(totChol)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp+Diab, family = binomial, data = data_na)
gam.diaBP1 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay,smoker)+s(totChol)+s(sysBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp+Diab, family = binomial, data = data_na)
gam.BMI1 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp+Diab, family = binomial, data = data_na)
gam.heartRate1 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp+Diab, family = binomial, data = data_na)
gam.glucose1 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+sex+OnBPMeds+PrevStroke+Hyp+Diab, family = binomial, data = data_na)
gam.sex1 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+OnBPMeds+PrevStroke+Hyp+Diab, family = binomial, data = data_na)
gam.smoker1 <- gam(CHD_Risk~s(age)+education+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp+Diab, family = binomial, data = data_na)
gam.OnBPMeds1 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke+Hyp+Diab, family = binomial, data = data_na)
gam.PrevStroke1 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+Hyp+Diab, family = binomial, data = data_na)
gam.Hyp1 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Diab, family = binomial, data = data_na)
gam.Diab1 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp, family = binomial, data = data_na)Check for significance.
anova(gam.age1, gam.init, test = "Chisq")
anova(gam.education1, gam.init, test = "Chisq")
anova(gam.totChol1, gam.init, test = "Chisq")
anova(gam.sysBP1, gam.init, test = "Chisq")
anova(gam.diaBP1, gam.init, test = "Chisq")
anova(gam.BMI1, gam.init, test = "Chisq")
anova(gam.heartRate1, gam.init, test = "Chisq")
anova(gam.glucose1, gam.init, test = "Chisq")
anova(gam.sex1, gam.init, test = "Chisq")
anova(gam.smoker1, gam.init, test = "Chisq")
anova(gam.OnBPMeds1, gam.init, test = "Chisq")
anova(gam.PrevStroke1, gam.init, test = "Chisq")
anova(gam.Hyp1, gam.init, test = "Chisq")
anova(gam.Diab1, gam.init, test = "Chisq")First, remove Diab.
gam.init2 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp, family = binomial, data = data_na)
gam.age2 <- gam(CHD_Risk~education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp, family = binomial, data = data_na)
gam.education2 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp, family = binomial, data = data_na)
gam.totChol2 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp, family = binomial, data = data_na)
gam.sysBP2 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp, family = binomial, data = data_na)
gam.diaBP2 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp, family = binomial, data = data_na)
gam.BMI2 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp, family = binomial, data = data_na)
gam.heartRate2 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp, family = binomial, data = data_na)
gam.glucose2 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+sex+OnBPMeds+PrevStroke+Hyp, family = binomial, data = data_na)
gam.sex2 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+OnBPMeds+PrevStroke+Hyp, family = binomial, data = data_na)
gam.smoker2 <- gam(CHD_Risk~s(age)+education+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke+Hyp, family = binomial, data = data_na)
gam.OnBPMeds2 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke+Hyp, family = binomial, data = data_na)
gam.PrevStroke2 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+Hyp, family = binomial, data = data_na)
gam.Hyp2 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+OnBPMeds+PrevStroke, family = binomial, data = data_na)Check for significance.
anova(gam.age2, gam.init2, test = "Chisq")
anova(gam.education2, gam.init2, test = "Chisq")
anova(gam.totChol2, gam.init2, test = "Chisq")
anova(gam.sysBP2, gam.init2, test = "Chisq")
anova(gam.diaBP2, gam.init2, test = "Chisq")
anova(gam.BMI2, gam.init2, test = "Chisq")
anova(gam.heartRate2, gam.init2, test = "Chisq")
anova(gam.glucose2, gam.init2, test = "Chisq")
anova(gam.sex2, gam.init2, test = "Chisq")
anova(gam.smoker2, gam.init2, test = "Chisq")
anova(gam.OnBPMeds2, gam.init2, test = "Chisq")
anova(gam.PrevStroke2, gam.init2, test = "Chisq")
anova(gam.Hyp2, gam.init2, test = "Chisq")Remove OnBPMeds.
gam.init3 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke+Hyp, family = binomial, data = data_na)
gam.age3 <- gam(CHD_Risk~education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke+Hyp, family = binomial, data = data_na)
gam.education3 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke+Hyp, family = binomial, data = data_na)
gam.totChol3 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke+Hyp, family = binomial, data = data_na)
gam.sysBP3 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke+Hyp, family = binomial, data = data_na)
gam.diaBP3 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke+Hyp, family = binomial, data = data_na)
gam.BMI3 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(heartRate)+s(glucose)+sex+PrevStroke+Hyp, family = binomial, data = data_na)
gam.heartRate3 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(glucose)+sex+PrevStroke+Hyp, family = binomial, data = data_na)
gam.glucose3 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+sex+PrevStroke+Hyp, family = binomial, data = data_na)
gam.sex3 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+PrevStroke+Hyp, family = binomial, data = data_na)
gam.smoker3 <- gam(CHD_Risk~s(age)+education+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke+Hyp, family = binomial, data = data_na)
gam.PrevStroke3 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+Hyp, family = binomial, data = data_na)
gam.Hyp3 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)Check for significance.
anova(gam.age3, gam.init3, test = "Chisq")
anova(gam.education3, gam.init3, test = "Chisq")
anova(gam.totChol3, gam.init3, test = "Chisq")
anova(gam.sysBP3, gam.init3, test = "Chisq")
anova(gam.diaBP3, gam.init3, test = "Chisq")
anova(gam.BMI3, gam.init3, test = "Chisq")
anova(gam.heartRate3, gam.init3, test = "Chisq")
anova(gam.glucose3, gam.init3, test = "Chisq")
anova(gam.sex3, gam.init3, test = "Chisq")
anova(gam.smoker3, gam.init3, test = "Chisq")
anova(gam.PrevStroke3, gam.init3, test = "Chisq")
anova(gam.Hyp3, gam.init3, test = "Chisq")Remove Hyp.
gam.init4 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.age4 <- gam(CHD_Risk~education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.education4 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.totChol4 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.sysBP4 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.diaBP4 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.BMI4 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.heartRate4 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.glucose4 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+sex+PrevStroke, family = binomial, data = data_na)
gam.sex4 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+PrevStroke, family = binomial, data = data_na)
gam.smoker4 <- gam(CHD_Risk~s(age)+education+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.PrevStroke4 <- gam(CHD_Risk~s(age)+education+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex, family = binomial, data = data_na)Check for significance.
anova(gam.age4, gam.init4, test = "Chisq")
anova(gam.education4, gam.init4, test = "Chisq")
anova(gam.totChol4, gam.init4, test = "Chisq")
anova(gam.sysBP4, gam.init4, test = "Chisq")
anova(gam.diaBP4, gam.init4, test = "Chisq")
anova(gam.BMI4, gam.init4, test = "Chisq")
anova(gam.heartRate4, gam.init4, test = "Chisq")
anova(gam.glucose4, gam.init4, test = "Chisq")
anova(gam.sex4, gam.init4, test = "Chisq")
anova(gam.smoker4, gam.init4, test = "Chisq")
anova(gam.PrevStroke4, gam.init4, test = "Chisq")Remove education.
gam.init5 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.age5 <- gam(CHD_Risk~s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.totChol5 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.sysBP5 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.diaBP5 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.BMI5 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.heartRate5 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.glucose5 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+sex+PrevStroke, family = binomial, data = data_na)
gam.sex5 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+PrevStroke, family = binomial, data = data_na)
gam.smoker5 <- gam(CHD_Risk~s(age)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.PrevStroke5 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(BMI)+s(heartRate)+s(glucose)+sex, family = binomial, data = data_na)Check for significance.
anova(gam.age5, gam.init5, test = "Chisq")
anova(gam.totChol5, gam.init5, test = "Chisq")
anova(gam.sysBP5, gam.init5, test = "Chisq")
anova(gam.diaBP5, gam.init5, test = "Chisq")
anova(gam.BMI5, gam.init5, test = "Chisq")
anova(gam.heartRate5, gam.init5, test = "Chisq")
anova(gam.glucose5, gam.init5, test = "Chisq")
anova(gam.sex5, gam.init5, test = "Chisq")
anova(gam.smoker5, gam.init5, test = "Chisq")
anova(gam.PrevStroke5, gam.init5, test = "Chisq")
# remove BMIRemove BMI.
gam.init6 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.age6 <- gam(CHD_Risk~s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.totChol6 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(sysBP)+s(diaBP)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.sysBP6 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(diaBP)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.diaBP6 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.heartRate6 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.glucose6 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(heartRate)+sex+PrevStroke, family = binomial, data = data_na)
gam.sex6 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(heartRate)+s(glucose)+PrevStroke, family = binomial, data = data_na)
gam.smoker6 <- gam(CHD_Risk~s(age)+s(totChol)+s(sysBP)+s(diaBP)+s(heartRate)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.PrevStroke6 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(heartRate)+s(glucose)+sex, family = binomial, data = data_na)Check for significance.
anova(gam.age6, gam.init6, test = "Chisq")
anova(gam.totChol6, gam.init6, test = "Chisq")
anova(gam.sysBP6, gam.init6, test = "Chisq")
anova(gam.diaBP6, gam.init6, test = "Chisq")
anova(gam.heartRate6, gam.init6, test = "Chisq")
anova(gam.glucose6, gam.init6, test = "Chisq")
anova(gam.sex6, gam.init6, test = "Chisq")
anova(gam.smoker6, gam.init6, test = "Chisq")
anova(gam.PrevStroke6, gam.init6, test = "Chisq")Remove heartrate
gam.init7 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.age7 <- gam(CHD_Risk~s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.totChol7 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(sysBP)+s(diaBP)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.sysBP7 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(diaBP)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.diaBP7 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.glucose7 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+sex+PrevStroke, family = binomial, data = data_na)
gam.sex7 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(glucose)+PrevStroke, family = binomial, data = data_na)
gam.smoker7 <- gam(CHD_Risk~s(age)+s(totChol)+s(sysBP)+s(diaBP)+s(glucose)+sex+PrevStroke, family = binomial, data = data_na)
gam.PrevStroke7 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(glucose)+sex, family = binomial, data = data_na)Check for Significance
anova(gam.age7, gam.init7, test = "Chisq")
anova(gam.totChol7, gam.init7, test = "Chisq")
anova(gam.sysBP7, gam.init7, test = "Chisq")
anova(gam.diaBP7, gam.init7, test = "Chisq")
anova(gam.glucose7, gam.init7, test = "Chisq")
anova(gam.sex7, gam.init7, test = "Chisq")
anova(gam.smoker7, gam.init7, test = "Chisq")
anova(gam.PrevStroke7, gam.init7, test = "Chisq")Remove PrevStroke.
gam.init8 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(glucose)+sex, family = binomial, data = data_na)
gam.age8 <- gam(CHD_Risk~s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(glucose)+sex, family = binomial, data = data_na)
gam.totChol8 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(sysBP)+s(diaBP)+s(glucose)+sex, family = binomial, data = data_na)
gam.sysBP8 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(diaBP)+s(glucose)+sex, family = binomial, data = data_na)
gam.diaBP8 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(glucose)+sex, family = binomial, data = data_na)
gam.glucose8 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+sex, family = binomial, data = data_na)
gam.sex8 <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)+s(sysBP)+s(diaBP)+s(glucose), family = binomial, data = data_na)
gam.smoker8 <- gam(CHD_Risk~s(age)+s(totChol)+s(sysBP)+s(diaBP)+s(glucose)+sex, family = binomial, data = data_na)Check for significance.
anova(gam.age8, gam.init8, test = "Chisq")
anova(gam.totChol8, gam.init8, test = "Chisq")
anova(gam.sysBP8, gam.init8, test = "Chisq")
anova(gam.diaBP8, gam.init8, test = "Chisq")
anova(gam.glucose8, gam.init8, test = "Chisq")
anova(gam.sex8, gam.init8, test = "Chisq")
anova(gam.smoker8, gam.init8, test = "Chisq")final.glm = glm(CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose + totChol, family = binomial, data = data_na)
gam.final <- gam(CHD_Risk~s(age)+s(cigsPerDay, smoker)+s(totChol)
+s(sysBP)+s(diaBP)+s(glucose)+sex,
family = binomial, data = data_na)
anova(final.glm, gam.final, test = "Chi")## Analysis of Deviance Table
##
## Model 1: CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay + glucose +
## totChol
## Model 2: CHD_Risk ~ s(age) + s(cigsPerDay, smoker) + s(totChol) + s(sysBP) +
## s(diaBP) + s(glucose) + sex
## Resid. Df Resid. Dev Df Deviance Pr(>Chi)
## 1 4231 3220.9
## 2 4221 3196.4 10.044 24.552 0.006415 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
summary(final.glm)##
## Call:
## glm(formula = CHD_Risk ~ 1 + age + sysBP + sex + smoker:cigsPerDay +
## glucose + totChol, family = binomial, data = data_na)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -2.0190 -0.5956 -0.4366 -0.2947 2.8407
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -8.845557 0.437862 -20.202 < 2e-16 ***
## age 0.064062 0.005962 10.745 < 2e-16 ***
## sysBP 0.016894 0.002006 8.421 < 2e-16 ***
## sexmale 0.510457 0.098074 5.205 1.94e-07 ***
## glucose 0.007577 0.001636 4.631 3.64e-06 ***
## totChol 0.001829 0.001023 1.788 0.0737 .
## smoker:cigsPerDay 0.010476 0.001928 5.435 5.49e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 3611.5 on 4237 degrees of freedom
## Residual deviance: 3220.9 on 4231 degrees of freedom
## AIC: 3234.9
##
## Number of Fisher Scoring iterations: 5
summary(gam.final)##
## Family: binomial
## Link function: logit
##
## Formula:
## CHD_Risk ~ s(age) + s(cigsPerDay, smoker) + s(totChol) + s(sysBP) +
## s(diaBP) + s(glucose) + sex
##
## Parametric coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -2.21037 0.07344 -30.097 < 2e-16 ***
## sexmale 0.51992 0.09935 5.233 1.67e-07 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Approximate significance of smooth terms:
## edf Ref.df Chi.sq p-value
## s(age) 2.405 3.028 109.411 < 2e-16 ***
## s(cigsPerDay,smoker) 2.818 3.404 28.270 6.94e-06 ***
## s(totChol) 1.006 1.012 3.501 0.0631 .
## s(sysBP) 1.000 1.000 26.196 3.09e-07 ***
## s(diaBP) 5.424 6.542 11.215 0.0956 .
## s(glucose) 2.390 2.989 22.080 6.85e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## R-sq.(adj) = 0.105 Deviance explained = 11.5%
## UBRE = -0.23774 Scale est. = 1 n = 4238