Multiple Sclerosis Lesion Segmentation Using Longitudinal Normalization and Convolutional Recurrent Neural Networks
Implementation in PyTorch of the paper Multiple Sclerosis Lesion Segmentation Using Longitudinal Normalization and Convolutional Recurrent Neural Networks and, more exactly, of the master thesis, pages 257-272 with the same name, for segmenting MS lesions from longitudinal multimodal MRI data.
Main script: cross_validation/cross_validation_3D_unet_convLSTM.py
Main model name: UNet_ConvLSTM_3D_alt_bidirectional (To be found in ms_segmentation/architectures/unet_c_gru.py). This is the actual CNN that combines the U-Net with the convolutional bidirectional LSTM.
Documentation and clean up in progress (slow progress). In script names CS is for cross-sectional and L for Longitudinal. Paths have to be corrected.
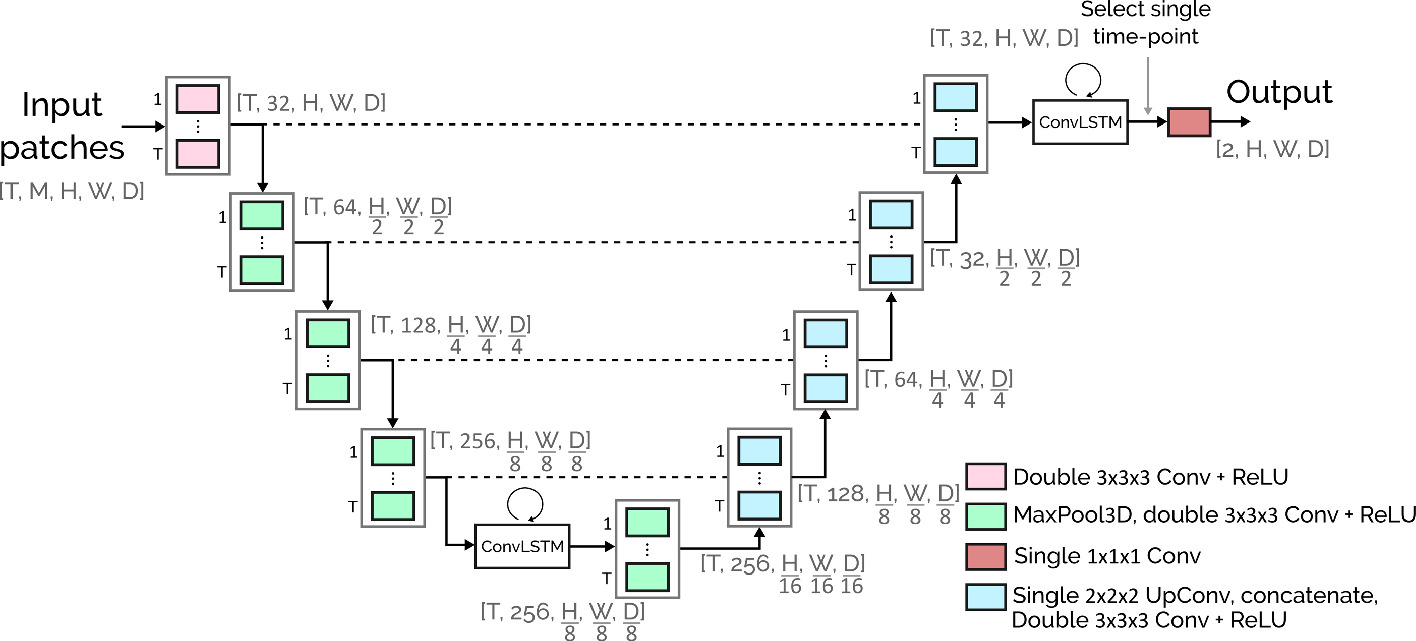
Architecture:
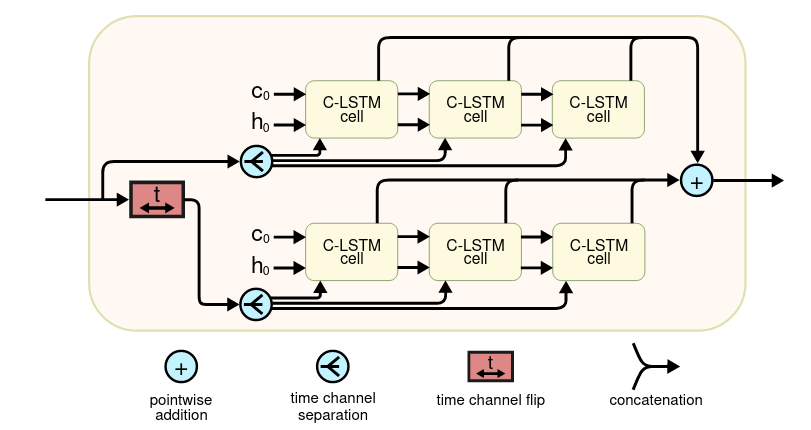
The architecture is a combination between the traditional U-Net and the convolutional LSTM, in its bidirectional version.
Each bidirectional block processes the patches of different time-points in both directions.
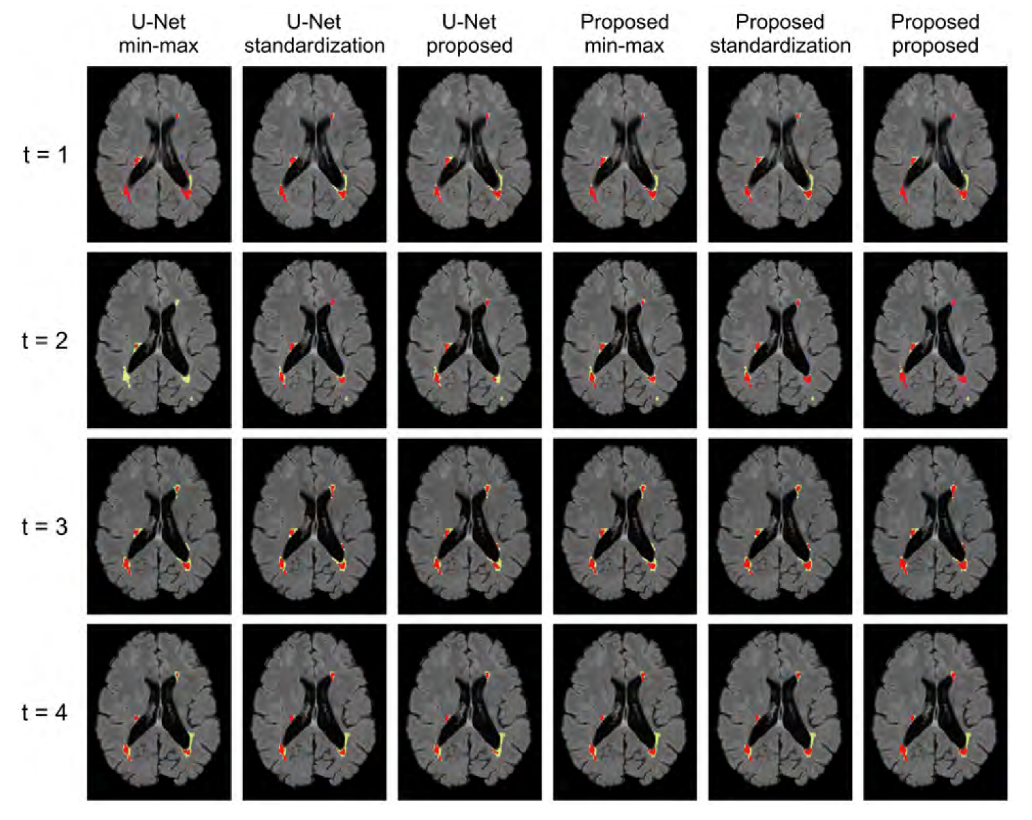
Here is an example of segmentation
Cite this work as:
Tascon-Morales, S., Hoffmann, S., Treiber, M., Mensing, D., Oliver, A., Guenther, M., & Gregori, J. (2020). Multiple Sclerosis Lesion Segmentation Using Longitudinal Normalization and Convolutional Recurrent Neural Networks. In Machine Learning in Clinical Neuroimaging and Radiogenomics in Neuro-oncology (pp. 148-158). Springer, Cham.