# Author : Amir Shokri
# github link : https://github.com/amirshnll/dermatology
# dataset link : http://archive.ics.uci.edu/ml/datasets/Dermatology
# email : amirsh.nll@gmail.comimport pandas as pd
from sklearn.preprocessing import MinMaxScaler
from sklearn.decomposition import PCA
from sklearn.model_selection import train_test_split
from sklearn.metrics import f1_score
from sklearn.metrics import accuracy_scoredata = pd.read_csv('dermatology_data.csv', header=None)data = data.replace(to_replace="?", method='ffill')data.describe()
<style scoped>
.dataframe tbody tr th:only-of-type {
vertical-align: middle;
}
</style>
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 34 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 366.000000 | 366.000000 | 366.000000 | 366.000000 | 366.000000 | 366.000000 | 366.000000 | 366.000000 | 366.000000 | 366.000000 | ... | 366.000000 | 366.000000 | 366.000000 | 366.000000 | 366.000000 | 366.000000 | 366.000000 | 366.000000 | 366.000000 | 366.000000 |
| mean | 2.068306 | 1.795082 | 1.549180 | 1.366120 | 0.633880 | 0.448087 | 0.166667 | 0.377049 | 0.614754 | 0.519126 | ... | 0.393443 | 0.464481 | 0.456284 | 0.953552 | 0.453552 | 0.103825 | 0.114754 | 1.866120 | 0.554645 | 2.803279 |
| std | 0.664753 | 0.701527 | 0.907525 | 1.138299 | 0.908016 | 0.957327 | 0.570588 | 0.834147 | 0.982979 | 0.905639 | ... | 0.849406 | 0.864899 | 0.954873 | 1.130172 | 0.954744 | 0.450433 | 0.488723 | 0.726108 | 1.105908 | 1.597803 |
| min | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 1.000000 |
| 25% | 2.000000 | 1.000000 | 1.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 1.000000 | 0.000000 | 1.000000 |
| 50% | 2.000000 | 2.000000 | 2.000000 | 1.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 2.000000 | 0.000000 | 3.000000 |
| 75% | 2.000000 | 2.000000 | 2.000000 | 2.000000 | 1.000000 | 0.000000 | 0.000000 | 0.000000 | 1.000000 | 1.000000 | ... | 0.000000 | 1.000000 | 0.000000 | 2.000000 | 0.000000 | 0.000000 | 0.000000 | 2.000000 | 0.000000 | 4.000000 |
| max | 3.000000 | 3.000000 | 3.000000 | 3.000000 | 3.000000 | 3.000000 | 3.000000 | 3.000000 | 3.000000 | 3.000000 | ... | 3.000000 | 3.000000 | 3.000000 | 3.000000 | 3.000000 | 3.000000 | 3.000000 | 3.000000 | 3.000000 | 6.000000 |
8 rows × 34 columns
properties = data[data.columns[:34]]
target = data[data.columns[34]]
scaler = MinMaxScaler()
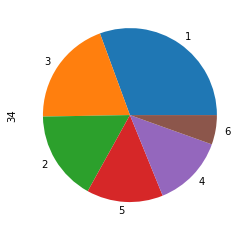
scaled_x = scaler.fit_transform(properties)target.value_counts().plot.pie()<matplotlib.axes._subplots.AxesSubplot at 0x23b944ca7f0>
pca = PCA(n_components=15)
reduced_x = pca.fit_transform(scaled_x)X_train, X_test, y_train, y_test = train_test_split(reduced_x, target, test_size=0.3, random_state=0)from sklearn.naive_bayes import GaussianNB
from sklearn.neural_network import MLPClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.linear_model import LogisticRegressiongnb = GaussianNB()
mlp = MLPClassifier(hidden_layer_sizes=(100, 100))
knn = KNeighborsClassifier(n_neighbors=5)
dt = DecisionTreeClassifier()
regressor = LogisticRegression()gnb.fit(X_train, y_train)
y_predgnb = gnb.predict(X_test)
mlp.fit(X_train, y_train)
y_predmlp = mlp.predict(X_test)
knn.fit(X_train, y_train)
y_predknn = knn.predict(X_test)
dt.fit(X_train, y_train)
y_preddt = dt.predict(X_test)
regressor.fit(X_train, y_train)
y_predregressor = regressor.predict(X_test)C:\Users\Amirshnll\anaconda3\lib\site-packages\sklearn\neural_network\_multilayer_perceptron.py:582: ConvergenceWarning: Stochastic Optimizer: Maximum iterations (200) reached and the optimization hasn't converged yet.
warnings.warn(
print('gnb f1: ', f1_score(y_test, y_predgnb, average='micro'))
print('gnb accuracy: ', accuracy_score(y_test, y_predgnb))
print('mlp f1: ', f1_score(y_test, y_predmlp, average='micro'))
print('mlp accuracy: ', accuracy_score(y_test, y_predmlp))
print('knn f1: ', f1_score(y_test, y_predgnb, average='micro'))
print('knn accuracy: ', accuracy_score(y_test, y_predknn))
print('decision tree f1: ', f1_score(y_test, y_predgnb, average='micro'))
print('decision tree accuracy: ', accuracy_score(y_test, y_preddt))
print('logistic regression f1: ', f1_score(y_test, y_predgnb, average='micro'))
print('logistic regression accuracy: ', accuracy_score(y_test, y_predregressor))gnb f1: 0.9727272727272728
gnb accuracy: 0.9727272727272728
mlp f1: 1.0
mlp accuracy: 1.0
knn f1: 0.9727272727272728
knn accuracy: 0.9818181818181818
decision tree f1: 0.9727272727272728
decision tree accuracy: 0.9090909090909091
logistic regression f1: 0.9727272727272728
logistic regression accuracy: 0.990909090909091