# Author : Amir Shokri
# github link : https://github.com/amirshnll/Guitar-Chords-finger-positions
# dataset link : http://archive.ics.uci.edu/ml/datasets/Guitar+Chords+finger+positions
# email : amirsh.nll@gmail.com
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from pandas.plotting import scatter_matrix
chord= pd.read_csv('chord-fingers.csv')
chord.head()
<style scoped>
.dataframe tbody tr th:only-of-type {
vertical-align: middle;
}
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
</style>
|
CHORD_ROOT |
CHORD_TYPE |
CHORD_STRUCTURE |
NOTE_NAMES |
chord1 |
chord2 |
FINGER_POSITIONS |
| 64 |
56 |
61 |
66 |
71 |
82 |
81 |
1 |
| 68 |
57 |
61 |
65 |
71 |
85 |
85 |
1 |
| 63 |
60 |
60 |
67 |
76 |
85 |
84 |
1 |
| 61 |
60 |
68 |
62 |
77 |
90 |
80 |
1 |
| 63 |
65 |
60 |
63 |
77 |
81 |
87 |
1 |
#chord.tail()
#chord.shape
#chord[:7]
chord.info()
#chord.columns
#chord['A'].unique()
#chord['B'].unique()
#chd['C'].unique()
#chord['D'].unique()
#chord['E'].unique()
#chord['F'].unique()
#chord['G'].unique()
#chord['ROOM'].unique()
#chord['A'].value_counts()
#chord['B'].value_counts()
#chord['C'].value_counts()
#chord['D'].value_counts()
#chord['E'].value_counts()
#chord['F'].value_counts()
#chord['G'].value_counts()
#chord['ROOM'].value_counts()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 2000 entries, 64 to 59
Data columns (total 7 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 CHORD_ROOT 2000 non-null int64
1 CHORD_TYPE 2000 non-null int64
2 CHORD_STRUCTURE 2000 non-null int64
3 NOTE_NAMES 2000 non-null int64
4 chord1 2000 non-null int64
5 chord2 2000 non-null int64
6 FINGER_POSITIONS 2000 non-null int64
dtypes: int64(7)
memory usage: 125.0 KB
chord.describe()
chord.hist(bins=50 , figsize=(20,15))
plt.show()
train_set,test_set=train_test_split(chord,test_size=0.2,random_state=42)
test_set.shape
data=train_set.copy()
#data.head(42)
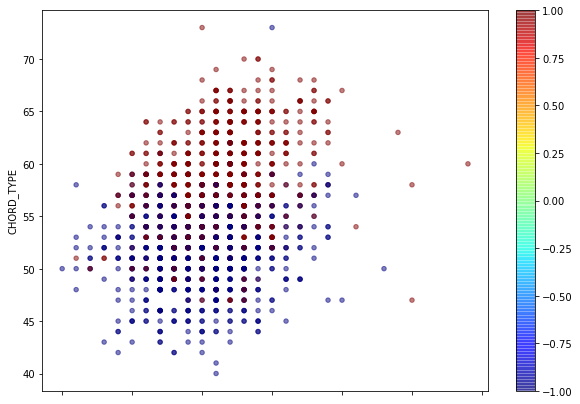
#standard correlation coefficient
data.plot(kind="scatter",x="CHORD_ROOT",y="CHORD_TYPE",
# s=data["B"]/2,label="",
c=data["FINGER_POSITIONS"],cmap=plt.get_cmap("jet"),
figsize=(10,7),alpha=0.5)
corr_matrix=data.corr()
corr_matrix["FINGER_POSITIONS"].sort_values(ascending=False)
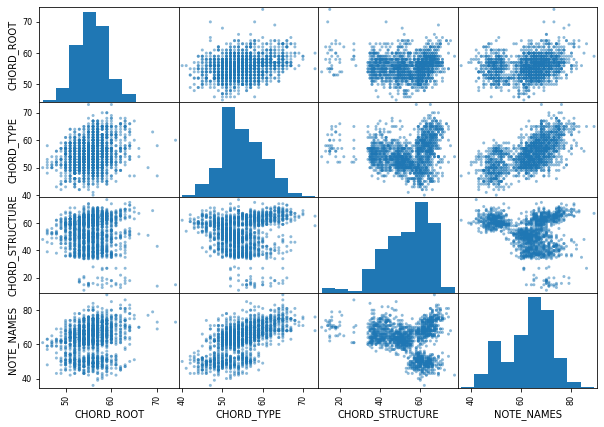
#scatter_matrix
feature=["CHORD_ROOT","CHORD_TYPE","CHORD_STRUCTURE","NOTE_NAMES"]
scatter_matrix(data[feature],figsize=(10,7))
plt.show()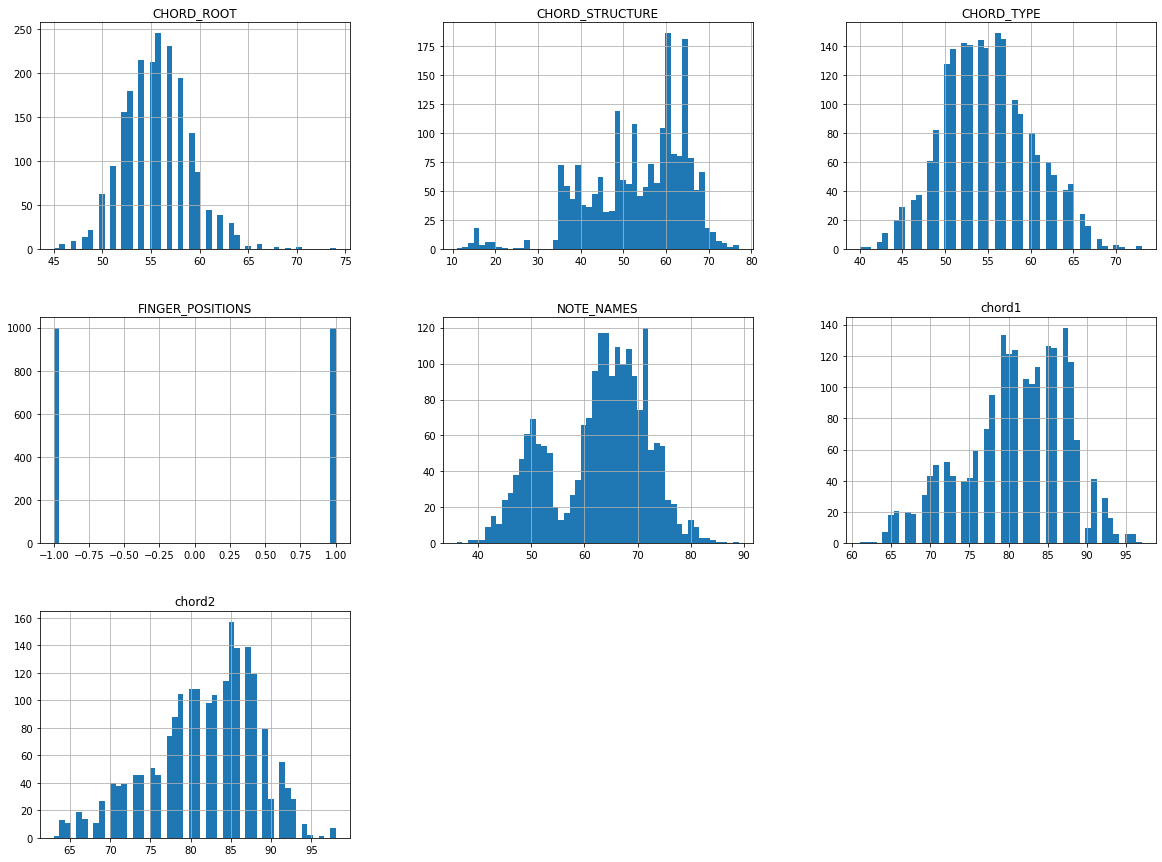
y=data.FINGER_POSITIONS
x_data=data.drop(columns=['FINGER_POSITIONS'])
print(x_data)
CHORD_ROOT CHORD_TYPE CHORD_STRUCTURE NOTE_NAMES chord1 chord2
37 54 47 36 63 70 70
58 59 59 65 65 82 94
42 54 58 41 63 75 78
37 56 57 39 61 72 74
64 57 63 59 68 82 83
.. ... ... ... ... ... ...
51 57 51 51 65 80 80
51 59 51 48 67 79 79
17 56 54 36 66 73 77
52 53 56 49 62 83 80
51 54 52 57 62 79 79
[1600 rows x 6 columns]
data = (x_data - np.min(x_data)) / (np.max(x_data) - np.min(x_data)).values
data.head()
<style scoped>
.dataframe tbody tr th:only-of-type {
vertical-align: middle;
}
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
</style>
|
CHORD_ROOT |
CHORD_TYPE |
CHORD_STRUCTURE |
NOTE_NAMES |
chord1 |
chord2 |
| 37 |
0.310345 |
0.212121 |
0.378788 |
0.509434 |
0.228571 |
0.200000 |
| 58 |
0.482759 |
0.575758 |
0.818182 |
0.547170 |
0.571429 |
0.885714 |
| 42 |
0.310345 |
0.545455 |
0.454545 |
0.509434 |
0.371429 |
0.428571 |
| 37 |
0.379310 |
0.515152 |
0.424242 |
0.471698 |
0.285714 |
0.314286 |
| 64 |
0.413793 |
0.696970 |
0.727273 |
0.603774 |
0.571429 |
0.571429 |
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(data,y,test_size = 0.2,random_state=150)
print("x_train: ",x_train.shape)
print("x_test: ",x_test.shape)
print("y_train: ",y_train.shape)
print("y_test: ",y_test.shape)x_train: (1280, 6)
x_test: (320, 6)
y_train: (1280,)
y_test: (320,)
# Import Decision Tree Classifier
from sklearn.tree import DecisionTreeClassifier
from sklearn import metrics
from sklearn import preprocessing
from sklearn.metrics import accuracy_score
clf = DecisionTreeClassifier()
clf = clf.fit(x_train,y_train)
from sklearn import tree
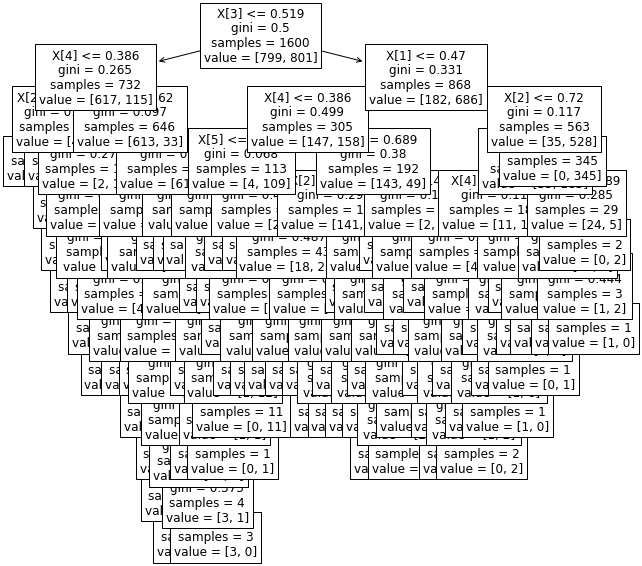
plt.figure(figsize=(10,10))
temp = tree.plot_tree(clf.fit(data,y), fontsize=12)
plt.show()
from sklearn.neighbors import KNeighborsClassifier
K = 5
knn = KNeighborsClassifier(n_neighbors=K)
knn.fit(x_train, y_train.ravel())
y_pred=knn.predict(x_test)
print("When K = {} neighnors , KNN test accuracy: {}".format(K, knn.score(x_test, y_test)))
print("When K = {} neighnors , KNN train accuracy: {}".format(K, knn.score(x_train, y_train)))
ran = np.arange(1,30)
train_list = []
test_list = []
for i,each in enumerate(ran):
knn = KNeighborsClassifier(n_neighbors=each)
knn.fit(x_train, y_train.ravel())
test_list.append(knn.score(x_test, y_test))
train_list.append(knn.score(x_train, y_train))
print("Best test score is {} , K = {}".format(np.max(test_list), test_list.index(np.max(test_list))+1))
print("Best train score is {} , K = {}".format(np.max(train_list), train_list.index(np.max(train_list))+1))When K = 5 neighnors , KNN test accuracy: 0.96875
When K = 5 neighnors , KNN train accuracy: 0.9875
Best test score is 0.971875 , K = 16
Best train score is 1.0 , K = 1
from sklearn.metrics import confusion_matrix as cm
cm(y_test, y_pred)
ax=sns.heatmap(cm(y_test, y_pred)/sum(sum(cm(y_test, y_pred))), annot=True)
b, t=ax.get_ylim()
ax.set_ylim(b+.5, t-.5)
plt.title('Confusion Matrix')
plt.ylabel('Truth')
plt.xlabel('Prediction')
plt.show();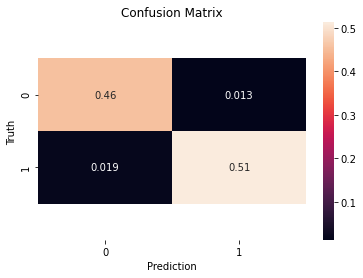
plt.figure(figsize=[15,10])
plt.plot(ran,test_list,label='Test Score')
plt.plot(ran,train_list,label = 'Train Score')
plt.xlabel('Number of Neighbers')
plt.ylabel('fav_number/retweet_count')
plt.xticks(ran)
plt.legend()
plt.show()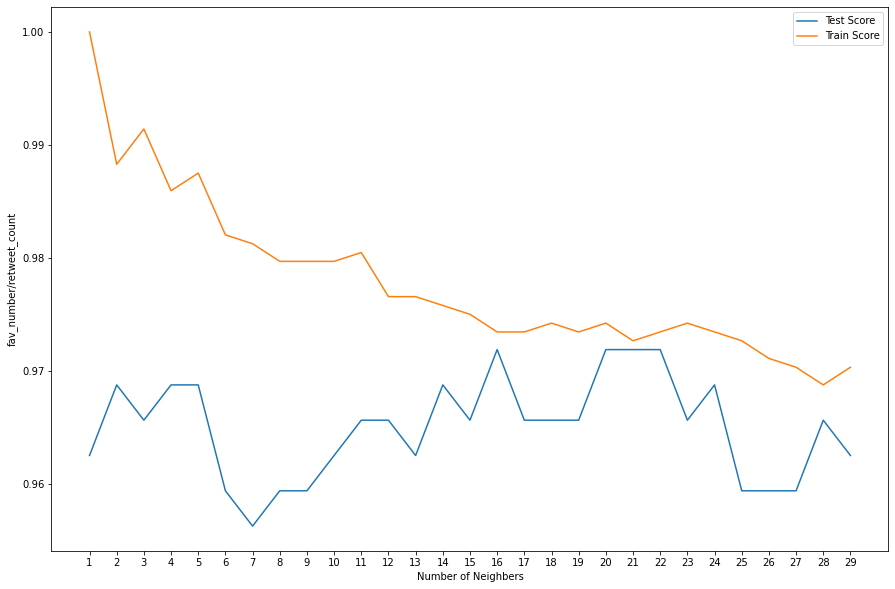
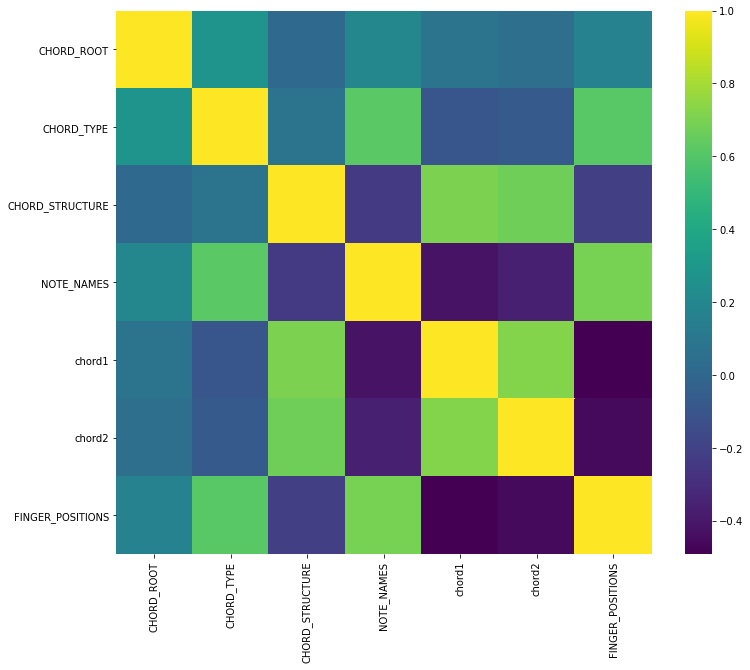
plt.figure(figsize=(12,10))
sns.heatmap(chord.corr(), cmap='viridis');
from sklearn.linear_model import Perceptron
clf = Perceptron(tol=1e-3, random_state=0)
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
from sklearn.metrics import classification_report
print(classification_report(y_test, clf.predict(x_test)))
print('Accuracy of logistic regression classifier on test set: {:.2f}'.format(clf.score(x_test, y_test))) precision recall f1-score support
-1 0.94 0.85 0.90 150
1 0.88 0.95 0.92 170
accuracy 0.91 320
macro avg 0.91 0.90 0.91 320
weighted avg 0.91 0.91 0.91 320
Accuracy of logistic regression classifier on test set: 0.91
from sklearn.metrics import classification_report, confusion_matrix
cm = confusion_matrix(y_test, y_pred)
fig, ax = plt.subplots(figsize=(8, 8))
ax.imshow(cm)
ax.grid(False)
ax.set_xlabel('Predicted outputs', fontsize=12, color='black')
ax.set_ylabel('Actual outputs', fontsize=8, color='black')
for i in range(2):
for j in range(2):
ax.text(j, i, cm[i, j], ha='center', va='center', color='black')
plt.show()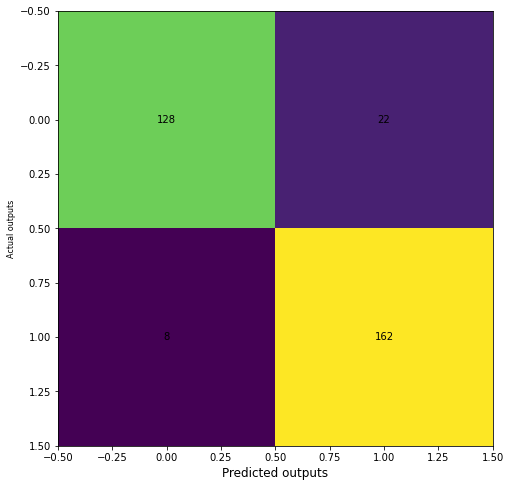
from sklearn.naive_bayes import GaussianNB
nb = GaussianNB()
nb.fit(x_train, y_train.ravel())
print("Naive Bayes test accuracy: ", nb.score(x_test, y_test))Naive Bayes test accuracy: 0.91875
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression(solver='lbfgs')
lr.fit(x_train, y_train.ravel())
y_pred = lr.predict(x_test)
from sklearn.metrics import classification_report
print(classification_report(y_test, lr.predict(x_test)))
print('Accuracy of logistic regression classifier on test set: {:.2f}'.format(lr.score(x_test, y_test))) precision recall f1-score support
-1 0.91 0.89 0.90 150
1 0.90 0.92 0.91 170
accuracy 0.91 320
macro avg 0.91 0.91 0.91 320
weighted avg 0.91 0.91 0.91 320
Accuracy of logistic regression classifier on test set: 0.91
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression(solver='lbfgs')
lr.fit(x_train, y_train.ravel())
y_pred = lr.predict(x_test)
from sklearn.metrics import classification_report
print(classification_report(y_test, lr.predict(x_test)))
print('Accuracy of logistic regression classifier on test set: {:.2f}'.format(lr.score(x_test, y_test))) precision recall f1-score support
-1 0.91 0.89 0.90 150
1 0.90 0.92 0.91 170
accuracy 0.91 320
macro avg 0.91 0.91 0.91 320
weighted avg 0.91 0.91 0.91 320
Accuracy of logistic regression classifier on test set: 0.91
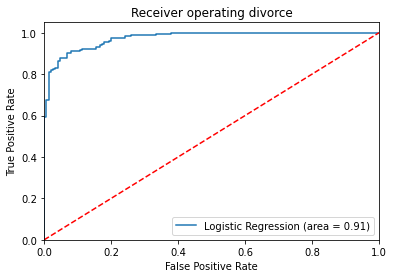
from sklearn.metrics import roc_auc_score
from sklearn.metrics import roc_curve
logit_roc_auc = roc_auc_score(y_test, lr.predict(x_test))
fpr, tpr, thresholds = roc_curve(y_test, lr.predict_proba(x_test)[:,1])
plt.figure()
plt.plot(fpr, tpr, label='Logistic Regression (area = %0.2f)' % logit_roc_auc)
plt.plot([0, 1], [0, 1],'r--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating divorce')
plt.legend(loc="lower right")
plt.savefig('Log_ROC')
plt.show()