An R package api to help run mosdepth and parse/visualise mosdepth output.
mosdepth is a commandline tool that quickly and efficiently calculates coverage from bam file inputs.
You can install the development version of mosdepth from GitHub with:
# install.packages("devtools")
devtools::install_github("selkamand/mosdepth")We start by loading the library
library(mosdepth)Then we point towards the mosdepth output directory
# Path to mosdepth output dir
mosdepth_dir = system.file("mosdepth_test_files", package="mosdepth")
# Create an object that keeps track of the output fils
mosdepth_output = mosdepth_collect_output_files(prefix = "E_coli", mosdepth_dir)
# Which output files do we have available? (not all options will produce the same set of files)
print(mosdepth_output)
#> summary ✔
#> global_distributions ✔
#> per_base_bed ✖
#> per_base_d4 ✔
#> regions_bed ✖
#> region_distributions ✖
#> quantized_bed ✖
#> thresholds_bed ✖Next we can run any mosdepth_plot_, mosdepth_tabulate_, or
mosdepth_calculate_ functions on this mosdepth output object. For
example we could plot cumulative coverage distributions
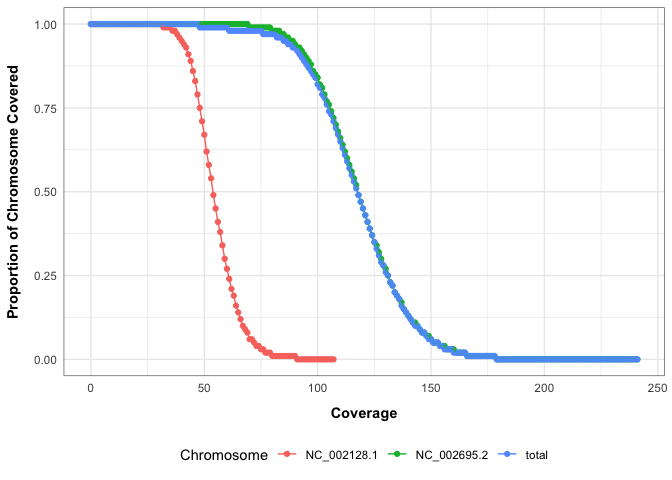
# Plot cumulative coverage distributions
mosdepth_plot_cumulative_coverage_distribution(mosdepth_output)Or get a dataframe containing summary metrics
# Get summary table
mosdepth_tabulate_summary(mosdepth_output)
#> chrom length bases mean min max
#> 1 NC_002695.2 5498578 649591221 118.14 0 280
#> 2 NC_002128.1 92721 5033884 54.29 18 109
#> 3 total 5591299 654625105 117.08 0 280