JSON Schema Witness Generation - Reproduction Package
This is a reproduction package for the article Witness Generation for JSON Schema by Lyes Attouche, Mohamed-Amine Baazizi, Dario Colazzo, Giorgio Ghelli, Carlo Sartiani, and Stefanie Scherzinger.
This reproduction package was created by Stefan Klessinger. It is provided as a Docker container.
Our original results, consisting of the table reporting our summary results as a csv and charts, are available in directory results.
Basic Reproduction
System Requirements
Docker and a Linux Kernel version compatible with Ubuntu 20.04 are required.
We successfully tested our reproduction package on both x86-64 and ARM64 (Apple M1) CPUs. Our experiments are configured to consume up to 10 GiB of RAM, in addition to RAM required by Docker and the host OS. Our testing systems had a minimum of 16 GB RAM.
Setting up the Docker Container
First, clone this repository with
git clone https://github.com/sdbs-uni-p/JSONSchemaWitnessGeneration.git.
To build the container, run docker build -t wg_repro . inside the root directory of this repository.
After building, start the container with docker run -it wg_repro. You may include the flag --name <name> (replacing <name> with a name of your choice) to identify the container more conveniently.
The contents of artifacts are copied into the container's home directory (/home/repro/). Paths described in this README are always relative to /home/repro/ (or artifacts, respectively) unless explicitly stated otherwise.
Reproduction in 3 Steps
The basic reproduction workflow inside the container is as follows:
- Run
./doAll.sh, executing all experiments, including chart generation and evaluation. On our test system with an Intel Xeon Silver 2.40GhZ CPU this took approximately 10 hours.- All results will be stored in
results, overwriting the original charts inresults/charts. Results for each dataset, including logs of errors and warnings, are stored in correspondingly named subdirectories, divided into satisfiable (-sat) and unsatisfiable (-unsat) schemas, if necessary. Additionally, results are summed up inresults.csvandevaluation.txt, the latter containing some additional comments.
- All results will be stored in
- Run
scripts/compare.sh, producing a diff-like comparison of the new results against our original results- By default, this script does not compare the system-specific runtimes. Use the optional parameter -t to enable comparison of runtime.
- Confirm successful reproduction:
- Inspect the output of
scripts/compare.sh -twhich produce an output showing different runtimes, like in the following screenshot (original results are shown in green, new results in red; use -c to disable colors):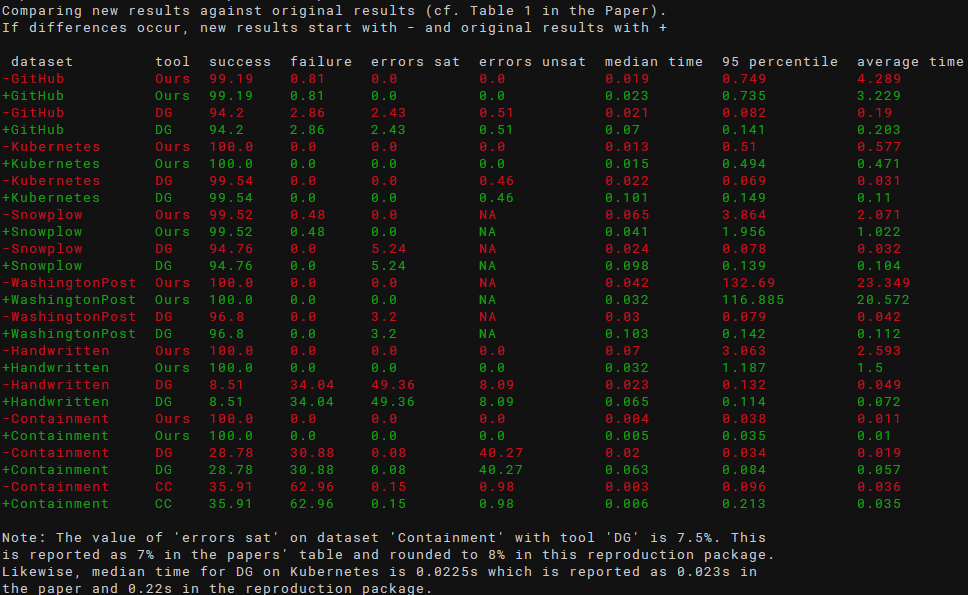
- Inspect the output of
scripts/compare.shwhich should show no differences and look similar to the following screenshot: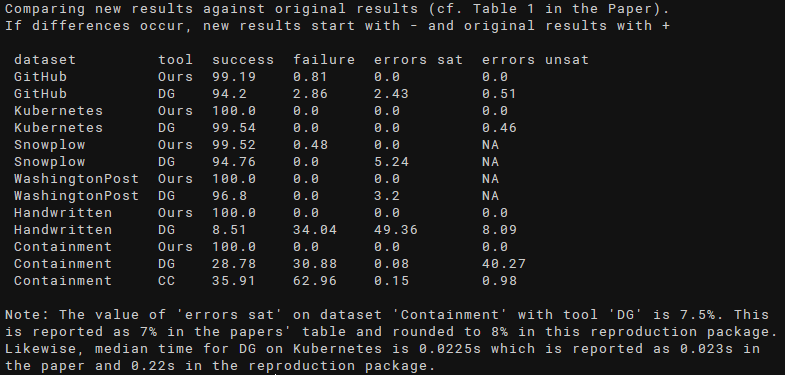
- Inspect
results/evaluation.txtfor additional insights into the evaluation, such as manual corrections we had to make, as also described in the paper. - To inspect the generated charts, they need to be copied to the host system - see section Moving Output to the Host System
- Inspect the output of
Advanced Topics
Artifact Availability
In this reproduction package, our own tool, as well as competitor tools are executed on the same datasets as in the accompanying paper. The datasets are stored in JSONAlgebra/JsonSchema_To_Algebra/expDataset. Datasets containing both, satisfiable and unsatisfiable, schemas are divided accordingly into subdirectories sat and unsat. In the GitHub dataset, we have different satisfiable datasets for jsongenerator (sat-dg) and our tool (sat), where different files are excluded as the tools had problems (e.g., exceeding the timeout) processing them. These files are considered as failures in our evaluation script. Likewise, the Snowplow dataset is divided into different datasets for jsongenerator (dg) and our tool (ours), the latter missing two schemas that exceeded the timeout during our experiments which are also counted as failures in our evaluation script.
We also provide the excluded GitHub and Snowplow schemas in the following directories (relative to JSONAlgebra/JsonSchema_To_Algebra/expDataset):
- github/excluded-ours contains 52 schemas that caused problems with our tool, grouped into subfolders by type of error.
- github/excluded-dg contains 10 schemas that caused problems with jsongenerator, grouped into subfolders by type of error.
- snowplow/excluded-ours contains two schemas on which our tool exceeded the specified timeout of one hour in our experiments.
These schemas are not processed by default when running our experiments. See section Executing Experiments on Specific Datasets on how to run experiments on these schemas.
The source code of our tool is available in JSONAlgebra.
Running Experiments
We provide a number of scripts for running our experiments inside the container.
To run all experiments including chart generation and evaluation, run ./doAll.sh. Note that this will take several hours (approximately 10 hours on our testing system).
There are additional scripts located in scripts for running individual experiments:
run-JSONAlgebra.shperforms experiments on all datasets with our tool. See the following sections for optional parameters.run-jsongenerator.shperforms experiments on all datasets with jsongenerator (DG).evalContainment.pyperforms and evaluates experiments on the containment dataset for jsonsubschema (CC). Opposed to the other tools, these experiments are performed from scratch with each execution and the results are not persisted.
Executing Experiments on Specific Datasets
To execute experiments on a specific dataset (e.g., the excluded datasets mentioned above), supply the optional parameter -i <dataset> to run-JSONAlgebra or run-jsongenerator, replacing <dataset> with the datasets path, relative to JSONAlgebra/JsonSchema_To_Algebra/expDataset.
For instance, to run experiments with our tool on github/excluded-ours/timeout, run ./run-JSONAlgebra -i github/excluded-ours/timeout. Results will be stored in a subdirectory of results, named like the dataset with slashes (/) replaced by dashes (-). Hence, in our example, results will be stored in results/github-excluded-ours-timeout.
Additional Settings
By default, run-JSONAlgebra.sh runs single-threaded without a timeout. The settings can be changed through parameters --timeout <ms>, allowing to specify the timeout in ms as an integer value.
In doAll.sh, a timeout of two hours per individual schema is set, to avoid excessively long runtimes. Our test dataset only contains schemas that were processed within an hour on our testing system. In case your system is considerably slower, please consider increasing the timeout.
Evaluating and Comparing Results
Evaluation is performed by evaluate.py in scripts. This script generates a text output describing results and comments on a few selected manual fixes we had to make, e.g., due to incorrect results produced by the third-party schema validator. In addition, a csv file, results.csv, mimicking Table 1 in our paper is stored in results. Experiments for jsonsubschema (CC) are created dynamically when running this script, with an execution time of approximately one minute on our systems.
When executing doAll.sh, the output of evaluate.py is stored in results/evaluation.txt.
To compare the computed results with the results stated in the paper, execute compare.sh in scripts. This compares results.csv, generated by evaluate.py, against original_results.csv, generating a diff-like output. compare.sh does not compare runtimes by default, since they will always differ between runs and takes two optional parameters:
-tenables comparison of runtimes-cdisables colored (red/green) output for reasons of visual accessibility
Generating Charts
To generate charts, execute create-charts.sh in charts inside the docker container. The generated charts are stored in results/charts, overwriting the original charts. When executing doAll.sh, charts are automatically generated and stored at the same location.
Moving Output to the Host System
All results are stored at /home/repro/results in the container. To copy the results to the host system, use docker cp <containerID>:/home/repro/results . having obtained the containerID using docker ps. If you use the --name flag, as explained above, you can replace the containerID with the chosen name.
Known Issues and Troubleshooting
Due to undeterministic behavior of Java datastructures in our implementation, a marginal deviation in results might occur. Specifically, we observed an issue with a single schema, o60883.json, in github/sat: Infrequently, processing of this schema does not terminate in reasonable time. We added a configurable timeout in doAll.sh, which causes a TimeoutException in these rare cases. Note that we do not recommend executing run-JSONAlgebra.sh without timeout enabled.
We further provide pre-compiled binary containers on Zenodo, in case external resources should become temporarily or permanently unavailable.
Should you run into other issues, we suggest the following approaches:
- Make sure that sufficient main memory is available on your system.
- Ensure that a sufficiently high memory limit is configured for the Docker container (e.g., in Docker Desktop)
- Run the experiments with our tool single-threaded (which is the default configuration).
- Increase the configured timeout in
doAll.sh.