Hedia Tnani
For the past 2 years, many groups have been working on creating APIs that the entire research community can leverage. There’s a publication A Review on Genomics APIs which shows some of the APIs used to retrieve data such as Google Genomics API and 23andMe API.
API stands for Application Programming Interface.
An API acts as interface between two applications (Client and sever).
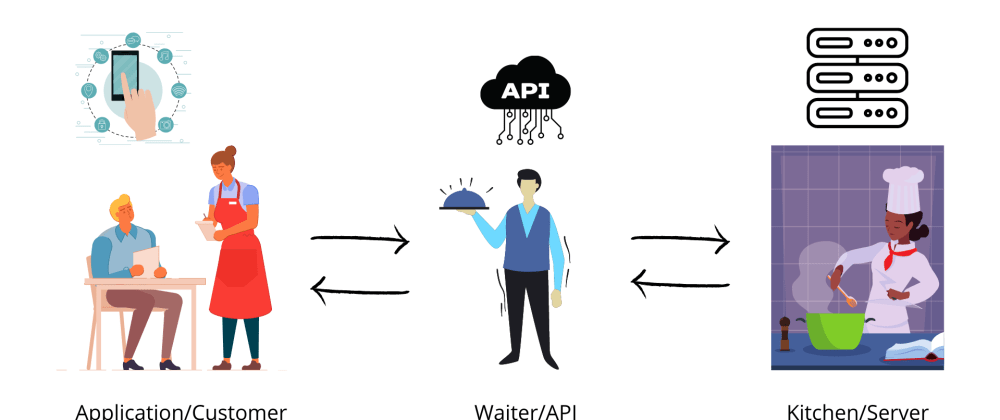
Let’s take a non technical example to make it to understand.
In terms of API, we can say that we as Customer(application) will send a request, the waiter(API) will take our request and send it to the chef(server). The chef(server) takes action according to our request and send it back to the waiter(API). This is how actually things work in the backend.
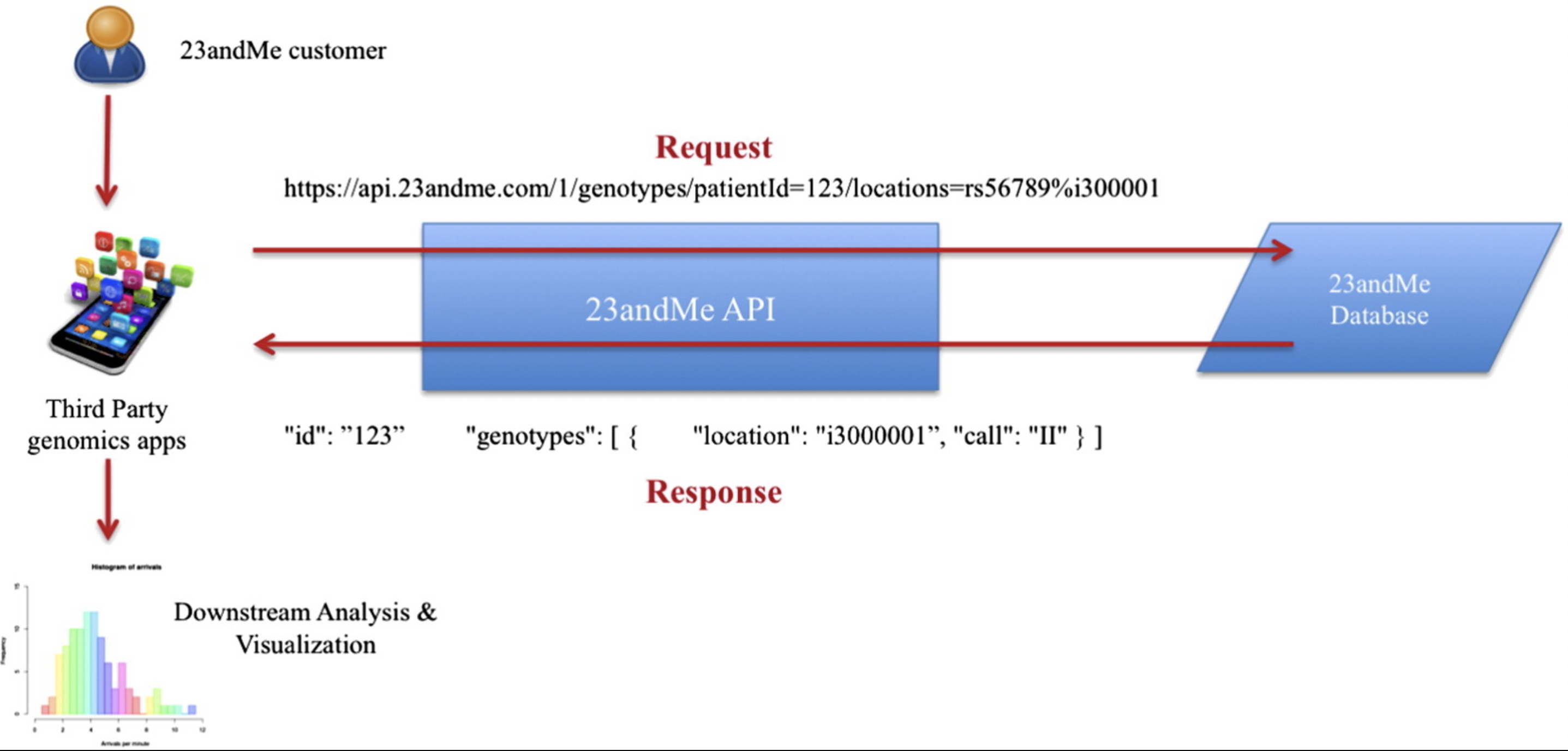
An example from the 23andMe API.
To work with APIs in R, we need to bring in two libraries
httr and jsonlite.
Every request starts with a URL.
The Uniform Resource Locator or URL is a string (of characters) that uniquely identifies a web resource. For example, “https://rest.ensembl.org” is the URL that identifies Ensembl’s homepage. The URL is the input of any API request. It consists of two parts: - The fixed part: base URL — the base URL is the main address of the API web server - The variable part: endpoint — an endpoint is a server route for retrieving specific data from an API.
API endpoints are the specific digital location where requests for information are sent by one program to retrieve the digital resource that exists there. Endpoints specify where APIs can access resources and help guarantee the proper functioning of the incorporated software. An API’s performance depends on its capacity to successfully communicate with API endpoints.
Example:
Let’s load the library(httr). The modify_url() allows us to combine
base URLs and endpoints.
library(httr)
# Preparing the URL
baseurl <- "https://rest.ensembl.org"
ext <- "/sequence/id/ENSG00000157764"
url <- modify_url(baseurl, path = ext)
url[1] "https://rest.ensembl.org/sequence/id/ENSG00000157764"
APIs use HTTP verbs for data requests. One of the most common is a
GET request. The GET request asks the API to retrieve a
resource, such as a database record or the contents of a file, and send
it to the client. To make a GET request we´ll be using the GET()
function from the httr package.
response = GET(url)
responseResponse [https://rest.ensembl.org/sequence/id/ENSG00000157764]
Date: 2023-02-24 11:44
Status: 200
Content-Type: application/json
Size: 206 kB
The API response contains, among other information, the query URL and date, the response status, and the content type and size. It might also include the content itself, but this part isn’t visible here.
The Content-Type of the API response is an application/json.
JSON stands for JavaScript Object Notation.
Most APIs return data in a JavaScript Object Notation (JSON) format. The JSON format is beneficial because
1) it’s a plain text file, and 2) it doesn’t need to be structured in a tabular data frame (i.e. is can store ‘non-rectangular’ or unstructured text easily).
When making queries, it’s entirely possible that the output isn’t the right type or that something will go wrong . Therefore, it’s important to track these errors. Web servers return status codes every time they receive an API request. A status code reports what happened with a request.
Let´s check the status code of our request:
http_status(response)$category
[1] "Success"
$reason
[1] "OK"
$message
[1] "Success: (200) OK"
The Status-Code element in a server response, is a 3-digit integer where the first digit of the Status-Code defines the class of response and the last two digits do not have any categorization role. There are 5 values for the first digit:
| S.N. | Code and Description |
|---|---|
| 1 | 1xx: Informational It means the request has been received and the process is continuing. |
| 2 | 2xx: Success It means the action was successfully received, understood, and accepted. |
| 3 | 3xx: Redirection It means further action must be taken in order to complete the request. |
| 4 | 4xx: Client Error It means the request contains incorrect syntax or cannot be fulfilled. |
| 5 | 5xx: Server Error It means the server failed to fulfill an apparently valid request. |
The httr package provides us with several functions to track these
errors.
- The
http_error(response)checks if something went wrong and returns a logical value.
http_error(response)[1] FALSE
- The
status_code(response)returns the status code.
status_code(response)[1] 200
- The
http_type(response)returns the format of the content, so we can see if it is what we expected.
http_type(response)[1] "application/json"
When something goes wrong and http_error(response) returns TRUE, we
stop the function execution using the stop() function.
if(http_error(response)){
status_code(404)
stop("Something went wrong.")}Another useful function is stop_for_status(response) which converts
http errors to R errors or warnings - these should always be used
whenever you’re creating requests inside a function, so that the user
knows why a request has failed. If request was successful, the response
(invisibly). Otherwise, raised a classed http error or warning, as
generated by http_condition() . Other helpful functions are
warn_for_status() and message_for_status().
One very useful function to use is content which extract content from
a request. For that we use content() from the httr package to
extract this content.
library(httr)
# automatically parses JSON
APIResult = content(response)
#APIResult$seqSo here we sent an API request to the webserver, and it replied with the sequence(data).
# add code to extract content here
json_text <- content(response, as="text")
#json_textLet’s add an encoding.
json_text <-content(response, as = "text", encoding = "UTF-8")
#json_textWe can provide the content_type in the GET().
Let’s get all microarrays available for a given species. In R, we can
parse this JSON string and convert it into a DataFrame using the
fromJSON() from the jsonlite package. If the output of an endpoint
contains a list, R will happily get all items in a list without the need
for any loops.
library(httr)
library(jsonlite)
library(xml2)
library(tidyverse)── Attaching packages ─────────────────────────────────────── tidyverse 1.3.2 ──
✔ ggplot2 3.4.0 ✔ purrr 0.3.5
✔ tibble 3.1.8 ✔ dplyr 1.0.10
✔ tidyr 1.2.1 ✔ stringr 1.4.1
✔ readr 2.1.3 ✔ forcats 0.5.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ purrr::flatten() masks jsonlite::flatten()
✖ dplyr::lag() masks stats::lag()
baseURL <- "https://rest.ensembl.org"
ext <- "/regulatory/species/homo_sapiens/microarray?"
r <- GET(paste(baseURL, ext, sep = ""), content_type("application/json"))
stop_for_status(r)
class(content(r)) # list[1] "list"
#fromJSON(content(r, "text", encoding = "UTF-8"))
#content(r, "text", encoding = "UTF-8")We can add the desired type of output: raw, text or
parsed. content attempts to automatically figure out which
one is most appropriate, based on the content-type.
identical(content(r,"parsed"), content(r))[1] TRUE
The fromJSON needs as an argument a JSON string, URL or file. To make
it a JSON string we convert it to text.
content(r, "text", encoding = "UTF-8")[1] "[{\"description\":null,\"array\":\"OneArray\",\"format\":\"EXPRESSION\",\"type\":\"OLIGO\",\"vendor\":\"PHALANX\"},{\"format\":\"EXPRESSION\",\"description\":null,\"array\":\"CODELINK\",\"vendor\":\"CODELINK\",\"type\":\"OLIGO\"},{\"format\":\"EXPRESSION\",\"array\":\"HumanWG_6_V3\",\"description\":null,\"vendor\":\"ILLUMINA\",\"type\":\"OLIGO\"},{\"description\":null,\"array\":\"GPL6848\",\"format\":\"EXPRESSION\",\"type\":\"OLIGO\",\"vendor\":\"AGILENT\"},{\"type\":\"OLIGO\",\"vendor\":\"ILLUMINA\",\"array\":\"HumanMethylation450\",\"description\":null,\"format\":\"METHYLATION\"},{\"type\":\"OLIGO\",\"vendor\":\"ILLUMINA\",\"format\":\"EXPRESSION\",\"array\":\"HumanRef-8_V3\",\"description\":null},{\"type\":\"OLIGO\",\"vendor\":\"AGILENT\",\"format\":\"EXPRESSION\",\"description\":null,\"array\":\"WholeGenome\"},{\"vendor\":\"AGILENT\",\"type\":\"OLIGO\",\"array\":\"CGH_44b\",\"description\":null,\"format\":\"CGH\"},{\"format\":\"EXPRESSION\",\"array\":\"SurePrint_G3_GE_8x60k\",\"description\":null,\"type\":\"OLIGO\",\"vendor\":\"AGILENT\"},{\"format\":\"EXPRESSION\",\"description\":null,\"array\":\"HC-G110\",\"vendor\":\"AFFY\",\"type\":\"OLIGO\"},{\"array\":\"SurePrint_G3_GE_8x60k_v2\",\"description\":null,\"format\":\"EXPRESSION\",\"vendor\":\"AGILENT\",\"type\":\"OLIGO\"},{\"format\":\"EXPRESSION\",\"array\":\"HG-Focus\",\"description\":null,\"type\":\"OLIGO\",\"vendor\":\"AFFY\"},{\"description\":null,\"array\":\"WholeGenome_4x44k_v1\",\"format\":\"EXPRESSION\",\"type\":\"OLIGO\",\"vendor\":\"AGILENT\"},{\"description\":null,\"array\":\"WholeGenome_4x44k_v2\",\"format\":\"EXPRESSION\",\"type\":\"OLIGO\",\"vendor\":\"AGILENT\"},{\"format\":\"EXPRESSION\",\"description\":null,\"array\":\"GPL26966\",\"type\":\"OLIGO\",\"vendor\":\"AGILENT\"},{\"type\":\"OLIGO\",\"vendor\":\"AFFY\",\"format\":\"EXPRESSION\",\"array\":\"HG-U133A_2\",\"description\":null},{\"vendor\":\"AGILENT\",\"type\":\"OLIGO\",\"format\":\"EXPRESSION\",\"description\":null,\"array\":\"GPL19072\"},{\"type\":\"OLIGO\",\"vendor\":\"ILLUMINA\",\"format\":\"METHYLATION\",\"description\":null,\"array\":\"HumanMethylation27\"},{\"description\":null,\"array\":\"HG-U133B\",\"format\":\"EXPRESSION\",\"vendor\":\"AFFY\",\"type\":\"OLIGO\"},{\"format\":\"EXPRESSION\",\"description\":\"Human Transcriptome Array 2.0\",\"array\":\"HTA-2_0\",\"type\":\"OLIGO\",\"vendor\":\"AFFY\"},{\"array\":\"HG-U133_Plus_2\",\"description\":null,\"format\":\"EXPRESSION\",\"vendor\":\"AFFY\",\"type\":\"OLIGO\"},{\"format\":\"EXPRESSION\",\"array\":\"HG_U95Av2\",\"description\":null,\"type\":\"OLIGO\",\"vendor\":\"AFFY\"},{\"format\":\"EXPRESSION\",\"array\":\"HG-U95B\",\"description\":null,\"type\":\"OLIGO\",\"vendor\":\"AFFY\"},{\"format\":\"EXPRESSION\",\"array\":\"HG-U95C\",\"description\":null,\"vendor\":\"AFFY\",\"type\":\"OLIGO\"},{\"description\":null,\"array\":\"HG-U95D\",\"format\":\"EXPRESSION\",\"type\":\"OLIGO\",\"vendor\":\"AFFY\"},{\"array\":\"HG-U95E\",\"description\":null,\"format\":\"EXPRESSION\",\"vendor\":\"AFFY\",\"type\":\"OLIGO\"},{\"type\":\"OLIGO\",\"vendor\":\"AFFY\",\"format\":\"EXPRESSION\",\"array\":\"HG_U95A\",\"description\":null},{\"vendor\":\"AFFY\",\"type\":\"OLIGO\",\"format\":\"EXPRESSION\",\"array\":\"HuGeneFL\",\"description\":null},{\"format\":\"EXPRESSION\",\"array\":\"U133_X3P\",\"description\":null,\"vendor\":\"AFFY\",\"type\":\"OLIGO\"},{\"format\":\"EXPRESSION\",\"description\":null,\"array\":\"PrimeView\",\"vendor\":\"AFFY\",\"type\":\"OLIGO\"},{\"vendor\":\"AFFY\",\"type\":\"OLIGO\",\"format\":\"EXPRESSION\",\"array\":\"HT_HG-U133_Plus_PM\",\"description\":null},{\"type\":\"OLIGO\",\"vendor\":\"AFFY\",\"format\":\"EXPRESSION\",\"array\":\"HuEx-1_0-st-v2\",\"description\":null},{\"type\":\"OLIGO\",\"vendor\":\"AFFY\",\"format\":\"EXPRESSION\",\"description\":null,\"array\":\"HuGene-2_0-st-v1\"},{\"vendor\":\"AFFY\",\"type\":\"OLIGO\",\"format\":\"EXPRESSION\",\"description\":null,\"array\":\"HuGene-1_0-st-v1\"},{\"vendor\":\"AFFY\",\"type\":\"OLIGO\",\"array\":\"HuGene-2_1-st-v1\",\"description\":null,\"format\":\"EXPRESSION\"}]"
To convert it to a dataframe we use the fromJSON function.
fromJSON(content(r, "text", encoding = "UTF-8")) description array format type
1 <NA> OneArray EXPRESSION OLIGO
2 <NA> CODELINK EXPRESSION OLIGO
3 <NA> HumanWG_6_V3 EXPRESSION OLIGO
4 <NA> GPL6848 EXPRESSION OLIGO
5 <NA> HumanMethylation450 METHYLATION OLIGO
6 <NA> HumanRef-8_V3 EXPRESSION OLIGO
7 <NA> WholeGenome EXPRESSION OLIGO
8 <NA> CGH_44b CGH OLIGO
9 <NA> SurePrint_G3_GE_8x60k EXPRESSION OLIGO
10 <NA> HC-G110 EXPRESSION OLIGO
11 <NA> SurePrint_G3_GE_8x60k_v2 EXPRESSION OLIGO
12 <NA> HG-Focus EXPRESSION OLIGO
13 <NA> WholeGenome_4x44k_v1 EXPRESSION OLIGO
14 <NA> WholeGenome_4x44k_v2 EXPRESSION OLIGO
15 <NA> GPL26966 EXPRESSION OLIGO
16 <NA> HG-U133A_2 EXPRESSION OLIGO
17 <NA> GPL19072 EXPRESSION OLIGO
18 <NA> HumanMethylation27 METHYLATION OLIGO
19 <NA> HG-U133B EXPRESSION OLIGO
20 Human Transcriptome Array 2.0 HTA-2_0 EXPRESSION OLIGO
21 <NA> HG-U133_Plus_2 EXPRESSION OLIGO
22 <NA> HG_U95Av2 EXPRESSION OLIGO
23 <NA> HG-U95B EXPRESSION OLIGO
24 <NA> HG-U95C EXPRESSION OLIGO
25 <NA> HG-U95D EXPRESSION OLIGO
26 <NA> HG-U95E EXPRESSION OLIGO
27 <NA> HG_U95A EXPRESSION OLIGO
28 <NA> HuGeneFL EXPRESSION OLIGO
29 <NA> U133_X3P EXPRESSION OLIGO
30 <NA> PrimeView EXPRESSION OLIGO
31 <NA> HT_HG-U133_Plus_PM EXPRESSION OLIGO
32 <NA> HuEx-1_0-st-v2 EXPRESSION OLIGO
33 <NA> HuGene-2_0-st-v1 EXPRESSION OLIGO
34 <NA> HuGene-1_0-st-v1 EXPRESSION OLIGO
35 <NA> HuGene-2_1-st-v1 EXPRESSION OLIGO
vendor
1 PHALANX
2 CODELINK
3 ILLUMINA
4 AGILENT
5 ILLUMINA
6 ILLUMINA
7 AGILENT
8 AGILENT
9 AGILENT
10 AFFY
11 AGILENT
12 AFFY
13 AGILENT
14 AGILENT
15 AGILENT
16 AFFY
17 AGILENT
18 ILLUMINA
19 AFFY
20 AFFY
21 AFFY
22 AFFY
23 AFFY
24 AFFY
25 AFFY
26 AFFY
27 AFFY
28 AFFY
29 AFFY
30 AFFY
31 AFFY
32 AFFY
33 AFFY
34 AFFY
35 AFFY
Let´s convert it to a tibble
as_tibble(fromJSON(content(r, "text", encoding = "UTF-8")))# A tibble: 35 × 5
description array format type vendor
<chr> <chr> <chr> <chr> <chr>
1 <NA> OneArray EXPRESSION OLIGO PHALANX
2 <NA> CODELINK EXPRESSION OLIGO CODELINK
3 <NA> HumanWG_6_V3 EXPRESSION OLIGO ILLUMINA
4 <NA> GPL6848 EXPRESSION OLIGO AGILENT
5 <NA> HumanMethylation450 METHYLATION OLIGO ILLUMINA
6 <NA> HumanRef-8_V3 EXPRESSION OLIGO ILLUMINA
7 <NA> WholeGenome EXPRESSION OLIGO AGILENT
8 <NA> CGH_44b CGH OLIGO AGILENT
9 <NA> SurePrint_G3_GE_8x60k EXPRESSION OLIGO AGILENT
10 <NA> HC-G110 EXPRESSION OLIGO AFFY
# … with 25 more rows
Let´s make a function.
library(httr)
library(jsonlite)
get_microarrays <- function(baseURL, ext, content_type){
r <- GET(paste(baseURL, ext, sep = ""), accept(content_type))
stop_for_status(r)
if (content_type == 'application/json'){
return (fromJSON(content(r, "text", encoding = "UTF-8")))
} else {
return (content(r, "text", encoding = "UTF-8"))
}
}
baseURL <- "https://rest.ensembl.org"
ext <- "/regulatory/species/homo_sapiens/microarray?"
con <- "application/json"
get_microarrays(baseURL, ext, con) type vendor format array
1 OLIGO PHALANX EXPRESSION OneArray
2 OLIGO CODELINK EXPRESSION CODELINK
3 OLIGO ILLUMINA EXPRESSION HumanWG_6_V3
4 OLIGO AGILENT EXPRESSION GPL6848
5 OLIGO ILLUMINA METHYLATION HumanMethylation450
6 OLIGO ILLUMINA EXPRESSION HumanRef-8_V3
7 OLIGO AGILENT EXPRESSION WholeGenome
8 OLIGO AGILENT CGH CGH_44b
9 OLIGO AGILENT EXPRESSION SurePrint_G3_GE_8x60k
10 OLIGO AFFY EXPRESSION HC-G110
11 OLIGO AGILENT EXPRESSION SurePrint_G3_GE_8x60k_v2
12 OLIGO AFFY EXPRESSION HG-Focus
13 OLIGO AGILENT EXPRESSION WholeGenome_4x44k_v1
14 OLIGO AGILENT EXPRESSION WholeGenome_4x44k_v2
15 OLIGO AGILENT EXPRESSION GPL26966
16 OLIGO AFFY EXPRESSION HG-U133A_2
17 OLIGO AGILENT EXPRESSION GPL19072
18 OLIGO ILLUMINA METHYLATION HumanMethylation27
19 OLIGO AFFY EXPRESSION HG-U133B
20 OLIGO AFFY EXPRESSION HTA-2_0
21 OLIGO AFFY EXPRESSION HG-U133_Plus_2
22 OLIGO AFFY EXPRESSION HG_U95Av2
23 OLIGO AFFY EXPRESSION HG-U95B
24 OLIGO AFFY EXPRESSION HG-U95C
25 OLIGO AFFY EXPRESSION HG-U95D
26 OLIGO AFFY EXPRESSION HG-U95E
27 OLIGO AFFY EXPRESSION HG_U95A
28 OLIGO AFFY EXPRESSION HuGeneFL
29 OLIGO AFFY EXPRESSION U133_X3P
30 OLIGO AFFY EXPRESSION PrimeView
31 OLIGO AFFY EXPRESSION HT_HG-U133_Plus_PM
32 OLIGO AFFY EXPRESSION HuEx-1_0-st-v2
33 OLIGO AFFY EXPRESSION HuGene-2_0-st-v1
34 OLIGO AFFY EXPRESSION HuGene-1_0-st-v1
35 OLIGO AFFY EXPRESSION HuGene-2_1-st-v1
description
1 <NA>
2 <NA>
3 <NA>
4 <NA>
5 <NA>
6 <NA>
7 <NA>
8 <NA>
9 <NA>
10 <NA>
11 <NA>
12 <NA>
13 <NA>
14 <NA>
15 <NA>
16 <NA>
17 <NA>
18 <NA>
19 <NA>
20 Human Transcriptome Array 2.0
21 <NA>
22 <NA>
23 <NA>
24 <NA>
25 <NA>
26 <NA>
27 <NA>
28 <NA>
29 <NA>
30 <NA>
31 <NA>
32 <NA>
33 <NA>
34 <NA>
35 <NA>
Let’s get the mouse homologue of the human BRCA2. If you specify another content type (not json), the helper function will get you this as text.
gene <- "BRCA2"
# define the URL parameters
server <- "http://rest.ensembl.org/"
con <- "application/json"
request <- paste("homology/symbol/human/", gene, "?target_species=mouse", sep = "")
r <- GET(paste(server, request, sep = ""), accept(con))
stop_for_status(r)
result = fromJSON(content(r, "text", encoding = "UTF-8"))
result$data$id[1] "ENSG00000139618"
Let’s retrieve the Kegg database using the Kegg API.
library(httr)
library(jsonlite)
library(xml2)
server <- "https://rest.kegg.jp"
ext <- "/info/hsa"
r <- GET(paste(server, ext, sep = ""), content_type("application/json"))
content(r)[1] "T01001 Homo sapiens (human) KEGG Genes Database\nhsa Release 105.0+/02-24, Feb 23\n Kanehisa Laboratories\n 24,729 entries\n\nlinked db pathway\n brite\n module\n ko\n genome\n enzyme\n network\n disease\n drug\n ncbi-geneid\n ncbi-proteinid\n uniprot\n"
Let’s have a look at the list of human pathways in Kegg.
library(httr)
library(jsonlite)
library(xml2)
library(tidyverse)
server <- "https://rest.kegg.jp"
ext <- "/list/pathway/hsa"
r <- GET(paste(server, ext, sep = ""), content_type("application/json"))
txt <- content(r, "text")
df <- tibble(kegg_results = read_lines(txt))df %>% separate(., col=kegg_results, into=c('Pathway_number', 'Pathway_name'), sep='\t')# A tibble: 352 × 2
Pathway_number Pathway_name
<chr> <chr>
1 path:hsa00010 Glycolysis / Gluconeogenesis - Homo sapiens (human)
2 path:hsa00020 Citrate cycle (TCA cycle) - Homo sapiens (human)
3 path:hsa00030 Pentose phosphate pathway - Homo sapiens (human)
4 path:hsa00040 Pentose and glucuronate interconversions - Homo sapiens (huma…
5 path:hsa00051 Fructose and mannose metabolism - Homo sapiens (human)
6 path:hsa00052 Galactose metabolism - Homo sapiens (human)
7 path:hsa00053 Ascorbate and aldarate metabolism - Homo sapiens (human)
8 path:hsa00061 Fatty acid biosynthesis - Homo sapiens (human)
9 path:hsa00062 Fatty acid elongation - Homo sapiens (human)
10 path:hsa00071 Fatty acid degradation - Homo sapiens (human)
# … with 342 more rows
Let’s retrieve the amino acid sequence of a human gene in a fasta
format.
server <- "https://rest.kegg.jp"
ext <- "/get/hsa:10458/aaseq"
r <- GET(paste(server, ext, sep = ""), content_type("text/x-fasta"))
print(content(r))[1] ">hsa:10458 K05627 BAI1-associated protein 2 | (RefSeq) BAIAP2, BAP2, FLAF3, IRSP53, WAML; BAR/IMD domain containing adaptor protein 2 (A)\nMSLSRSEEMHRLTENVYKTIMEQFNPSLRNFIAMGKNYEKALAGVTYAAKGYFDALVKMG\nELASESQGSKELGDVLFQMAEVHRQIQNQLEEMLKSFHNELLTQLEQKVELDSRYLSAAL\nKKYQTEQRSKGDALDKCQAELKKLRKKSQGSKNPQKYSDKELQYIDAISNKQGELENYVS\nDGYKTALTEERRRFCFLVEKQCAVAKNSAAYHSKGKELLAQKLPLWQQACADPSKIPERA\nVQLMQQVASNGATLPSALSASKSNLVISDPIPGAKPLPVPPELAPFVGRMSAQESTPIMN\nGVTGPDGEDYSPWADRKAAQPKSLSPPQSQSKLSDSYSNTLPVRKSVTPKNSYATTENKT\nLPRSSSMAAGLERNGRMRVKAIFSHAAGDNSTLLSFKEGDLITLLVPEARDGWHYGESEK\nTKMRGWFPFSYTRVLDSDGSDRLHMSLQQGKSSSTGNLLDKDDLAIPPPDYGAASRAFPA\nQTASGFKQRPYSVAVPAFSQGLDDYGARSMSRNPFAHVQLKPTVTNDRCDLSAQGPEGRE\nHGDGSARTLAGR\n"
Let’s try the xml content_type.
server <- "https://rest.kegg.jp"
ext <- "/get/hsa00600/kgml"
r <- GET(paste(server, ext, sep = ""), content_type("text/xml"))
content(r,encoding = "UTF-8"){xml_document}
<pathway name="path:hsa00600" org="hsa" number="00600" title="Sphingolipid metabolism" image="https://www.kegg.jp/kegg/pathway/hsa/hsa00600.png" link="https://www.kegg.jp/kegg-bin/show_pathway?hsa00600">
[1] <entry id="128" name="hsa:5660 hsa:768239" type="gene" link="https://www ...
[2] <entry id="35" name="hsa:55331" type="gene" reaction="rn:R06518" link="h ...
[3] <entry id="36" name="ko:K05848" type="ortholog" reaction="rn:R06528" lin ...
[4] <entry id="38" name="hsa:130367 hsa:81537" type="gene" reaction="rn:R065 ...
[5] <entry id="39" name="hsa:130367 hsa:81537" type="gene" reaction="rn:R065 ...
[6] <entry id="40" name="hsa:130367 hsa:81537" type="gene" reaction="rn:R065 ...
[7] <entry id="41" name="hsa:427 hsa:56624" type="gene" reaction="rn:R06518" ...
[8] <entry id="42" name="hsa:427 hsa:56624" type="gene" reaction="rn:R06528" ...
[9] <entry id="43" name="hsa:10715 hsa:204219 hsa:253782 hsa:29956 hsa:79603 ...
[10] <entry id="44" name="hsa:123099 hsa:8560" type="gene" reaction="rn:R0651 ...
[11] <entry id="45" name="ko:K04709" type="ortholog" reaction="rn:R06527" lin ...
[12] <entry id="46" name="hsa:123099 hsa:8560" type="gene" reaction="rn:R0652 ...
[13] <entry id="47" name="hsa:123099 hsa:8560" type="gene" reaction="rn:R0652 ...
[14] <entry id="48" name="hsa:339221 hsa:55512 hsa:55627 hsa:6609 hsa:6610" t ...
[15] <entry id="49" name="hsa:10715 hsa:204219 hsa:253782 hsa:29956 hsa:79603 ...
[16] <entry id="51" name="ko:K26283" type="ortholog" reaction="rn:R01503" lin ...
[17] <entry id="52" name="ko:K01123" type="ortholog" reaction="rn:R02542" lin ...
[18] <entry id="53" name="hsa:64781" type="gene" reaction="rn:R01495" link="h ...
[19] <entry id="54" name="hsa:8611 hsa:8612 hsa:8613" type="gene" reaction="r ...
[20] <entry id="55" name="hsa:8611 hsa:8612 hsa:8613" type="gene" reaction="r ...
...
Let´s have a look at the 23andme API. Let’s get the list of accessions available on 23andMe platforms.
library(httr2)Attaching package: 'httr2'
The following object is masked from 'package:xml2':
url_parse
req <- request("https://api.23andme.com/3/accession/")
req<httr2_request>
GET https://api.23andme.com/3/accession/
Body: empty
We can see exactly what this request will send to the server with a dry run:
req %>% req_dry_run()GET /3/accession/ HTTP/1.1
Host: api.23andme.com
User-Agent: httr2/0.2.2 r-curl/4.3.3 libcurl/7.86.0
Accept: */*
Accept-Encoding: deflate, gzip
What's changed with httr?
-
The method has changed from GET to POST. POST is the standard method for sending data to a website, and is automatically used whenever you add a body. Use
req_method()to for a different method. -
There are two new headers:
Content-TypeandContent-Length. They tell the server how to interpret the body — it's encoded as JSON and is 15 bytes long. -
We have a body, consisting of some JSON.
To actually perform a request and fetch the response back from the
server, call req_perform()
resp <- req %>% req_perform()
resp<httr2_response>
GET https://api.23andme.com/3/accession/
Status: 200 OK
Content-Type: application/json
Body: In memory (1474 bytes)
Responses with status codes 4xx and 5xx are HTTP errors. httr2
automatically turns these into R errors. This is another important
difference to httr, which required that you explicitly call
httr::stop_for_status() to turn HTTP errors into R errors.
You can extract the body in various forms using the resp_body_*()
family of functions. Since this response returns JSON we can use
resp_body_json():
resp %>% resp_body_json() $data
$data[[1]]
$data[[1]]$id
[1] "NC_000001.10"
$data[[1]]$chromosome
[1] "1"
$data[[1]]$length
[1] 249250621
$data[[2]]
$data[[2]]$id
[1] "NC_000002.11"
$data[[2]]$chromosome
[1] "2"
$data[[2]]$length
[1] 243199373
$data[[3]]
$data[[3]]$id
[1] "NC_000003.11"
$data[[3]]$chromosome
[1] "3"
$data[[3]]$length
[1] 198022430
$data[[4]]
$data[[4]]$id
[1] "NC_000004.11"
$data[[4]]$chromosome
[1] "4"
$data[[4]]$length
[1] 191154276
$data[[5]]
$data[[5]]$id
[1] "NC_000005.9"
$data[[5]]$chromosome
[1] "5"
$data[[5]]$length
[1] 180915260
$data[[6]]
$data[[6]]$id
[1] "NC_000006.11"
$data[[6]]$chromosome
[1] "6"
$data[[6]]$length
[1] 171115067
$data[[7]]
$data[[7]]$id
[1] "NC_000007.13"
$data[[7]]$chromosome
[1] "7"
$data[[7]]$length
[1] 159138663
$data[[8]]
$data[[8]]$id
[1] "NC_000008.10"
$data[[8]]$chromosome
[1] "8"
$data[[8]]$length
[1] 146364022
$data[[9]]
$data[[9]]$id
[1] "NC_000009.11"
$data[[9]]$chromosome
[1] "9"
$data[[9]]$length
[1] 141213431
$data[[10]]
$data[[10]]$id
[1] "NC_000010.10"
$data[[10]]$chromosome
[1] "10"
$data[[10]]$length
[1] 135534747
$data[[11]]
$data[[11]]$id
[1] "NC_000011.9"
$data[[11]]$chromosome
[1] "11"
$data[[11]]$length
[1] 135006516
$data[[12]]
$data[[12]]$id
[1] "NC_000012.11"
$data[[12]]$chromosome
[1] "12"
$data[[12]]$length
[1] 133851895
$data[[13]]
$data[[13]]$id
[1] "NC_000013.10"
$data[[13]]$chromosome
[1] "13"
$data[[13]]$length
[1] 115169878
$data[[14]]
$data[[14]]$id
[1] "NC_000014.8"
$data[[14]]$chromosome
[1] "14"
$data[[14]]$length
[1] 107349540
$data[[15]]
$data[[15]]$id
[1] "NC_000015.9"
$data[[15]]$chromosome
[1] "15"
$data[[15]]$length
[1] 102531392
$data[[16]]
$data[[16]]$id
[1] "NC_000016.9"
$data[[16]]$chromosome
[1] "16"
$data[[16]]$length
[1] 90354753
$data[[17]]
$data[[17]]$id
[1] "NC_000017.10"
$data[[17]]$chromosome
[1] "17"
$data[[17]]$length
[1] 81195210
$data[[18]]
$data[[18]]$id
[1] "NC_000018.9"
$data[[18]]$chromosome
[1] "18"
$data[[18]]$length
[1] 78077248
$data[[19]]
$data[[19]]$id
[1] "NC_000019.9"
$data[[19]]$chromosome
[1] "19"
$data[[19]]$length
[1] 59128983
$data[[20]]
$data[[20]]$id
[1] "NC_000020.10"
$data[[20]]$chromosome
[1] "20"
$data[[20]]$length
[1] 63025520
$data[[21]]
$data[[21]]$id
[1] "NC_000021.8"
$data[[21]]$chromosome
[1] "21"
$data[[21]]$length
[1] 48129895
$data[[22]]
$data[[22]]$id
[1] "NC_000022.10"
$data[[22]]$chromosome
[1] "22"
$data[[22]]$length
[1] 51304566
$data[[23]]
$data[[23]]$id
[1] "NC_000023.10"
$data[[23]]$chromosome
[1] "X"
$data[[23]]$length
[1] 155270560
$data[[24]]
$data[[24]]$id
[1] "NC_000024.9"
$data[[24]]$chromosome
[1] "Y"
$data[[24]]$length
[1] 59373566
$data[[25]]
$data[[25]]$id
[1] "NC_012920.1"
$data[[25]]$chromosome
[1] "MT"
$data[[25]]$length
[1] 16569
$links
$links$`next`
NULL
https://www.w3schools.com/js/js_json_intro.asp
https://www.tutorialspoint.com/http/http_status_codes.htm
https://medium.com/geekculture/a-beginners-guide-to-apis-9aa7b1b2e172
https://httr2.r-lib.org/articles/httr2.html
https://httr2.r-lib.org/articles/wrapping-apis.html#faker-api
https://www.storybench.org/how-to-access-apis-in-r/