The pajengr package has been created to facilite the data manipulation
of trace files in the Paje Trace File Format. You provide a paje trace
file, and you get multiple tibbles (aka data frames) with information
about states, links, variables, events, and containers. These tables
have information that is very similar to the ones provided by the pj_dump tool.
Refer to the PajeNG dependencies before proceeding with the pajengr R
package installation.
Then, you need devtools package.
install.packages("devtools");Finally, install pajengr like this:
library(devtools);
install_github("schnorr/pajengr");As of now, the pajeng_read is the only exported function of the
pajengr package. This function returns a list with five named
elements: “container”, “state”, “variable”, “event”, and “link”. Each
element is a data.frame with the same columns as the output given by
the pj_dump tool. The code snippet below shows the basic usage of the
pajeng_read function, which takes the name of a file in the Paje file
format.
library(pajengr)
suppressMessages(library(tidyverse))
data <- pajeng_read("traces/simu-mardi.trace");
data$container %>% as_tibble()
data$state %>% as_tibble()
data$variable %>% as_tibble()
data$event %>% as_tibble()
data$link %>% as_tibble()# A tibble: 307 x 7 Nature Parent Type Start End Duration Container <fct> <fct> <fct> <dbl> <dbl> <dbl> <fct> 1 Container NULL 0 0 1205 1205 0 2 Container 0 L1 0 1205 1205 my_cluster_1 3 Container my_cluster_1 ROUTER 0 1205 1205 nodemy_cluster_1_router 4 Container my_cluster_1 LINK 0 1205 1205 my_cluster_1_link_32_UP 5 Container my_cluster_1 LINK 0 1205 1205 my_cluster_1_link_31_DOWN 6 Container my_cluster_1 LINK 0 1205 1205 my_cluster_1_link_31_UP 7 Container my_cluster_1 LINK 0 1205 1205 my_cluster_1_link_30_DOWN 8 Container my_cluster_1 LINK 0 1205 1205 my_cluster_1_link_30_UP 9 Container my_cluster_1 LINK 0 1205 1205 my_cluster_1_link_29_DOWN 10 Container my_cluster_1 LINK 0 1205 1205 my_cluster_1_link_29_UP # ... with 297 more rows # A tibble: 13,620 x 8 Nature Container Type Start End Duration Imbrication Value <fct> <fct> <fct> <dbl> <dbl> <dbl> <int> <fct> 1 State node32 PM 100 222 122 0 normal 2 State node32 PM 222 256 34 0 normal 3 State node32 PM 256 268 12 0 normal 4 State node32 PM 268 291 23 0 normal 5 State node32 PM 291 303 12 0 normal 6 State node32 PM 303 425 122 0 normal 7 State node32 PM 425 503 78 0 normal 8 State node32 PM 503 515 12 0 normal 9 State node32 PM 515 538 23 0 normal 10 State node32 PM 538 554 16 0 normal # ... with 13,610 more rows # A tibble: 507 x 7 Nature Container Type Start End Duration Value <fct> <fct> <fct> <dbl> <dbl> <dbl> <dbl> 1 Variable my_cluster_1_link_32_UP bandwidth 0 1205 1205 1.25e+8 2 Variable my_cluster_1_link_32_UP latency 0 1205 1205 5.00e-5 3 Variable my_cluster_1_link_31_DOWN bandwidth 0 1205 1205 1.25e+8 4 Variable my_cluster_1_link_31_DOWN latency 0 1205 1205 5.00e-5 5 Variable my_cluster_1_link_31_UP bandwidth 0 1205 1205 1.25e+8 6 Variable my_cluster_1_link_31_UP latency 0 1205 1205 5.00e-5 7 Variable my_cluster_1_link_30_DOWN bandwidth 0 1205 1205 1.25e+8 8 Variable my_cluster_1_link_30_DOWN latency 0 1205 1205 5.00e-5 9 Variable my_cluster_1_link_30_UP bandwidth 0 1205 1205 1.25e+8 10 Variable my_cluster_1_link_30_UP latency 0 1205 1205 5.00e-5 # ... with 497 more rows # A tibble: 0 x 5 # ... with 5 variables: Nature <fct>, Container <fct>, Type <fct>, Start <dbl>, # Value <fct> # A tibble: 405 x 10 Nature Container Type Start End Duration Value StartContainer EndContainer <fct> <fct> <fct> <dbl> <dbl> <dbl> <fct> <fct> <fct> 1 State my_clust… L1-L… 0 0 0 G my_cluster_1_… node21 2 State my_clust… L1-L… 0 0 0 G my_cluster_1_… node22 3 State my_clust… L1-L… 0 0 0 G my_cluster_1_… node23 4 State my_clust… L1-L… 0 0 0 G my_cluster_1_… node24 5 State my_clust… L1-L… 0 0 0 G my_cluster_1_… node25 6 State my_clust… L1-L… 0 0 0 G my_cluster_1_… node26 7 State my_clust… L1-L… 0 0 0 G my_cluster_1_… node27 8 State my_clust… L1-L… 0 0 0 G my_cluster_1_… node28 9 State my_clust… L1-L… 0 0 0 G my_cluster_1_… node29 10 State my_clust… L1-L… 0 0 0 G my_cluster_1_… node30 # ... with 395 more rows, and 1 more variable: Key <fct>
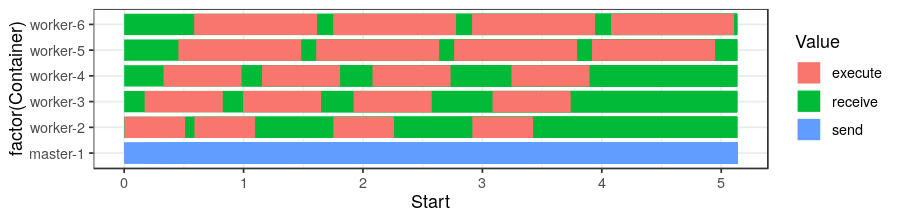
One might want to create a space/time view (without imbrication):
library(pajengr)
suppressMessages(library(tidyverse))
pajeng_read("traces/simgrid.trace")$state %>%
ggplot(aes(x=Start, xend=End, y=factor(Container),yend=factor(Container), color=Value)) +
theme_bw(base_size=18) +
geom_segment(size=10)Despite being available in the pj_dump tool, the features below are
currently unsupported in the pajengr package. They are expected to be
integrated in this package in the future. Open up an issue if some of
these is important for you.
- Use the flex parser (
--flex) - Export user fields (
--user-defined) - Dump ends at timestamp END (
--end=END) - Dump starts at timestamp START (instead of 0) (
--start=START) - No imbrication levels (push and pop become sets) (
--no-imbrication) - Support old field names in event definitions (
--no-strict) - Out of core execution (smallest memory footprint) (
--out-of-core) - Ignore incomplete links (not recommended) (
--ignore-incomplete-links)
Use the Issue tab or get in touch by e-mail with: