Image Quality Transfer (IQT) aims to bridge the technological gap that exists between bespoke and expensive experimental systems such as the Human Connectome Project (HCP) scanner and accessible commercial clinical systems using machine learning (ML). The technique learns mappings from low quality (e.g. clinical) to high quality (e.g. experimental) images exploiting the similarity of images across subjects, regions, modalities, and scales: image macro- and meso-structure is highly predictive of sub-voxel content. The mapping may then operate directly on low-quality images to estimate the corresponding high-quality images, or serve as a prior in an otherwise ill-posed image-reconstruction routine.
The current version provides a MATLAB implementation of IQT for super-resolution of diffusion tensor images (DTIs) using random forests (RFs).
To achieve the best quality, training should be done on datasets available from the HCP project. A demonstration script is provided, which also requires this data and can be accessed freely from here.
There are two parts to using the IQT software: application and training (which also includes a demonstration).
For each script you need to edit the Settings section to set the correct paths and select your parameters.
Pretrained random forest models for super-resolution that were trained following the paper (see Citation) are included along with this software. These can be used to super-resolve your DTI data.
| command | description |
|---|---|
compute_model_dti.m |
Compute DTI on your data and save in IQT compatible format. |
reconstruct_hires_dti.m |
Compute super-resolution DTI from your "low-quality" DTI. |
Pretrained trees can be found in the directory trees. These trees can perform x2 or x3 super-resolution with input patches of size 5x5x5.
By default, the reconstruction on the outer boundary of the brain requires a separate reconstruction method and is ignored. If you wish to perform boundary completion,
set construct_edge = 1 in the Settings. Reconstruction takes about 10 minutes (2 hours) for each subject with 8 trees without (with) boundary completion.
If you wish to train your own trees for your problem, you can use the following functions along with the recommended HCP data.
| command | description |
|---|---|
train_preprocess.m |
Creates the training data from the chosen subjects and generates paired patch libraries. |
train_rf.m |
Trains the chosen number of trees from the paired patch libraries. |
test_rf.m |
A demonstration that can be used to get a quick flavour of the IQT code. It is also useful to visualise the results of the training. It requires HCP data and automatically visualises the mean diffusivity (MD), fractional anisotropy (FA) and colour encoded directional map (CFA) of the predicted high-resolution DTI, and saves all in a MATLAB FIG file. |
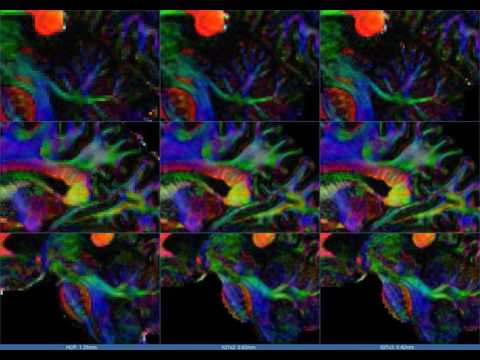
The figure below is a typical visualisation from test_rf.m and illustrates results of x3 super-resolution
with 3x3x3 input patch on subject 117324. Note that no boundary completion was performed here.
A script is provided to convert the super-resolved DTI from the IQT compatible format to standard 4D NIFTI format.
| command | description |
|---|---|
dti_from_IQT_format.m |
Convert DTI from the IQT compatible format to a 4D NIFTI. Default element ordering is MRtrix3 compatible (Dxx, Dyy, Dzz, Dxy, Dxz, Dyz). |
If you use this pipeline in your research, please cite:
@article{alexander2017image,
title={Image quality transfer and applications in diffusion MRI},
author={Daniel C. Alexander and Darko Zikic and Aurobrata Ghosh and Ryutaro Tanno and Viktor Wottschel and Jiaying Zhang and Enrico Kaden and Tim B Dyrby and Stamatios N Sotiropoulos and Hui Zhang and Antonio Criminisi},
journal={NeuroImage},
volume={152},
pages={283--298},
year={2017},
publisher={Elsevier}
}
NeuroImage paper: "Image quality transfer and applications in diffusion MRI" (link).
This YouTube video showcases x2 and x3 super-resolved DTI CFA maps computed from the original HCP data, going from 1.25mm to 0.63mm and 0.42mm. More details are in the video's description.
- The current version is limited to super-resolution of diffusion tensor images (DTIs).
- Boundary reconstruction can take some time (~2 hours per subject). It's best to try the pipeline first without this feature (
construct_edge = 0in theSettings).