This repository contains supplementary information to
Niklas E. Siedhoff1,§, Alexander-Maurice Illig1,§, Ulrich Schwaneberg1,2, Mehdi D. Davari1,*,
PyPEF – An Integrated Framework for Data-Driven Protein Engineering, J. Chem. Inf. Model. 2021, 61, 3463-3476
https://doi.org/10.1021/acs.jcim.1c00099
1Institute of Biotechnology, RWTH Aachen University, Worringer Weg 3, 52074 Aachen, Germany
2DWI-Leibniz Institute for Interactive Materials, Forckenbeckstraße 50, 52074 Aachen, Germany
*Corresponding author
§Equal contribution
a framework written in Python 3 for performing sequence-based machine learning-assisted protein engineering to predict a protein's fitness from its sequence.
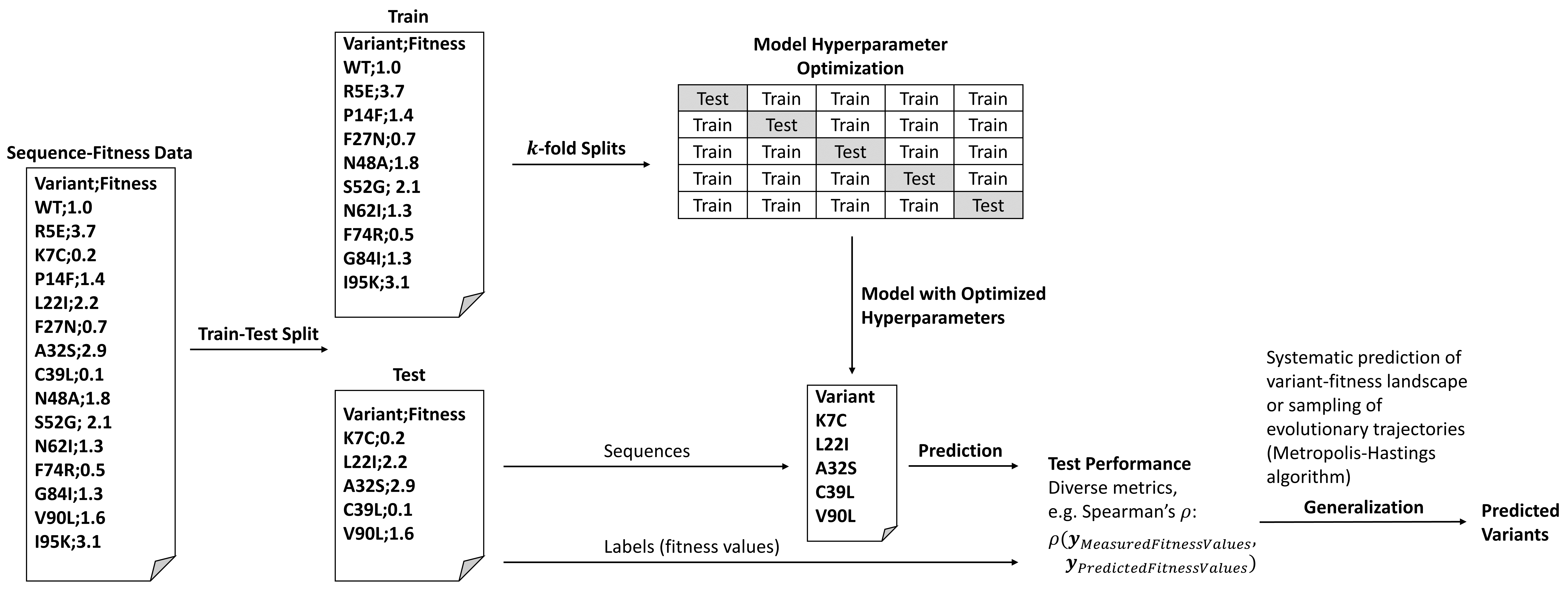
Protein engineering by rational or random approaches generates data that can aid the construction of self-learned sequence-function landscapes to predict beneficial variants by using probabilistic methods that can screen the unexplored sequence space with uncertainty in silico. Such predictive methods can be applied for increasing the success/effectivity of an engineering campaign while partly offering the prospect to reveal (higher-order) epistatic effects. Here we present an engineering framework termed PyPEF for assisting the supervised training and testing of regression models for predicting beneficial combinations of (identified) amino acid substitutions using machine learning algorithms from the scikit-learn package. As training input, the developed framework requires the variant sequences and the corresponding screening results (fitness labels) of the identified variants as CSV (or FASTA-like datasets following a self-defined convention). Using linear or nonlinear regression methods (partial least squares (PLS), support vector machines (SVR), random forest (RF), Ridge, LassoLars, and multilayer perceptron (MLP)-based regression), PyPEF trains on the given learning data while optimizing model hyperparameters (default: five-fold cross-validation) and can compute model performances on left-out test data. As sequences are encoded using amino acid descriptor sets taken from the AAindex database, finding the best index-dependent encoding for a specific test set can be seen as a hyperparameter search on the test set. Additional sequence- and system-specific encoding techniques will be added in future. Finally, the selected or best identified model can be used to perform directed evolution walks in silico (see Church-lab implementation or the reimplementation) or to predict natural diverse or recombinant sequences that subsequently are to be designed and validated in the wet-lab.
For detailed information, please refer to the above-mentioned publication and related Supporting Information.
The workflow procedure is explained in the Jupyter Notebook (.ipynb) protocol (see Tutorial section below and the ./workflow directory).
PyPEF was developed to be run from a command-line interface while python3 ./pypef/cli/run.py (when using the downloaded version of this repository) should be equal to pypef when installed with pip.
pypef --help
pypef mklsvs -w WT_SEQUENCE.FASTA -i VARIANT-FITNESS_DATA.CSV
pypef run -l LEARNING_SET.FASTA -v VALIDATION_SET.FASTA --regressor TYPE
pypef --show
pypef run -m MODEL12345 -f VALIDATION_SET.FASTA
pypef run -m MODEL12345 -p PREDICTION_SET.FASTA
pypef mkps -w WT_SEQUENCE.FASTA -i VARIANT-FITNESS_DATA.CSV --drecomb
pypef run -m MODEL12345 --pmult --drecomb
pypef directevo -m MODEL12345 --ywt WT_FITNESS -w WT_SEQUENCE.FASTA --usecsv -i VARIANT-FITNESS_DATA.CSV
Sample files for testing PyPEF routines are provided in the workflow/test_dataset directory, which is also used when running the Notebook tutorial. PyPEF's package dependencies are linked here. A small example of using the encoding API for sequence encoding and model validation is provided in the encoding_validation_api directory. Further, for designing your own API based on the PyPEF workflow, modules can be adapted from the source code provided in the pypef source directory. A quick installation of the PyPEF command line framework using PyPI (tested for Linux OS and Python 3.7–3.9) can be performed with:
pip install pypef
After successful installation, PyPEF should work by calling pypef in the shell, e.g.:
pypef --help
The detailed routine for setting up a new virtual environment using Anaconda, installing required Python packages for that environment, and running the Jupyter Notebook tutorial is given below.
As standard input files, PyPEF requires the target protein wild-type sequence in FASTA format and variant-fitness data in CSV format to create learning and validation split files that resemble the aligned FASTA format and additionally contain lines indicating the fitness of each corresponding variant (see sample files).
Before starting running the tutorial, it is a good idea to set-up a new Python environment using Anaconda, https://www.anaconda.com/, e.g. using Anaconda (Anaconda3-2020.11-Linux-x86_64.sh installer download) or Miniconda. Change to the download directory and run the installation, e.g. in Linux:
bash Anaconda3-2020.11-Linux-x86_64.sh
After accepting all steps, the conda setup should also be written to your ~/.bashrcfile, so that you can call anaconda typing conda.
Next, to download this repository click Code > Download ZIP and unzip the zipped file, e.g. with unzip PyPEF-main.zip, or just clone this repository using your bash shell to your local machine git clone https://github.com/Protein-Engineering-Framework/PyPEF.
To setup a new environment with conda you can either create the conda environment from the provided YAML file inside the PyPEF directory (cd PyPEF or cd PyPEF-main dependent on the downloaded file name):
conda env create --file pypef_environment.yml
or you can create a new environment yourself. You just need to specify the name of the environment and the Python version, e.g.:
conda create --name pypef python=3.7
To activate the environment you can define:
conda activate pypef
After activating the environment you can install required packages after changing the directory to the PyPEF directory (cd PyPEF or cd PyPEF-main) and install required packages with pip if you did not use the YAML file for creating the environment (when using conda packages will be installed in anaconda3/envs/pypef/lib/python3.7/site-packages):
python3 -m pip install -r requirements.txt
Note that the package Ray that we use for parallelization of the model validation routine does not yet fully support running on Windows and does not need to be used when running on a single core (no --parallelflag).
Now, after installing required packages, you should be able to directly run pypef in your preferred command-line interface (see running example).
To run the tutorial after installing required packages either from the conda YAML environment file, the TEXT requirement file, or after installing packages using the pip version of PyPEF, you have to open a Jupyter Notebook. If you have installed Anaconda, Jupyter Notebook and other commonly used packages for scientific computing and data science should be already installed in Python. If not, you can also install Jupyter via python3 -m pip install ipython jupyter. To use the pypef environment as kernel inside the Jupyter Notebook, you need to install the ipykernel:
python3 -m pip install ipykernel
python3 -m ipykernel install --user --name=pypef
Now change the directory to ./workflow (cd workflow) and run the .ipynb file:
jupyter-notebook
Copy the Notebook URL in your internet browser and select the Workflow_PyPEF.ipynb file to open it. Now you can select the pypef Python environment at the top Notebook menu: Kernel > Change kernel > pypef (otherwise you would use your default Python version as environment, i.e. you would have to install the required packages for this interpreter as well; for this case the installation of the prerequisite packages can also be done within the Notebook in provided code fields).
Good luck and have fun!
- Sequence encoding based on AAindex descriptor sets; e.g., sequence 'ACDEFGACDEFG' --> [0.61, 1.07, 0.46, 0.47, 2.02, 0.07, 0.61, 1.07, 0.46, 0.47, 2.02, 0.07] using AAindex https://www.genome.jp/entry/aaindex:ARGP820101 for encoding and without FFT (and 'ACDEFGACDEFG' --> [1.253E-32, 1.076E-03, 3.281E-02, 1.507E-01, 7.837E-34, 1.000E00, 7.787E-01, 1.521E-01] with FFT)
- to be implemented
- to be implemented
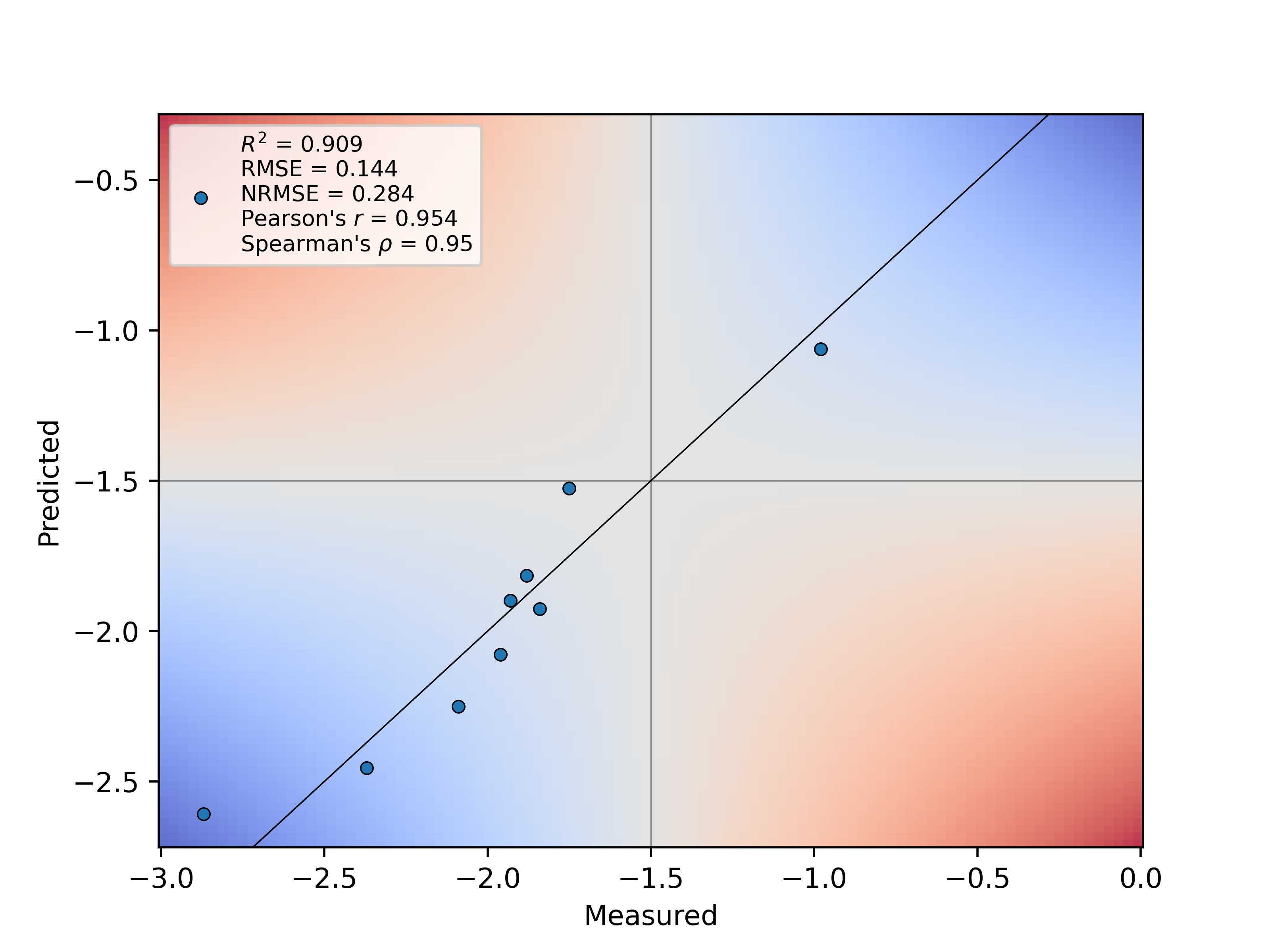
The following model hyperparameter ranges are tested during (k-fold) cross-validation for optimized model generalization:
| Regression model | Hyperparameter grid |
|---|---|
| PLS | N_components= {1, 2, 3, ..., 9} |
| RF | N_trees = {100, 250, 500, 1000}, max. features = {all features, all features, log2(all features)} |
| SVR | regularization param. = {2^0, 2^2, 2^4, 2^6, 2^8, 2^10, 2^12}, kernel coefficient = {1E−01, 1E−02, 1E−03, 1E−04, 1E−05} |
| MLP | single hidden layer size = {1, 2, ..., 12}, solver = {ADAM, L-BFGS}, initial learning rate = {0.001, 0.01, 0.1} |
| LassoLars | regularization param. = {1.000E-06, 1.322E-06, 1.748E-06, ..., 1.000E06} (numpy.logspace(-6, 6, 100)), to be implemented |
| Ridge | regularization param. = {1.000E-06, 1.322E-06, 1.748E-06, ..., 1.000E06} (numpy.logspace(-6, 6, 100)), to be implemented |