Code to pre-process whole body scans (DXA, MRI) for experiments in the paper 'Self-Supervised Multi-Modal Alignment For Whole Body Medical Imaging'. This code can also be used as pre-processing for other studies which rely on the UK Biobank DXA/Dixon MR datasets.
Please see our paper here. The full code release for this paper is available here. You are welcome to use this code either to reproduce the results of our experiments or for data preprocessing in your own research. If you do, please cite the following:
Windsor, R., Jamaludin, A., Kadir, T. ,Zisserman, A. "Self-Supervised Multi-Modal Alignment For Whole Body Medical Imaging" In: Proceedings of 24th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2021
bibtex:
@inproceedings{Windsor21SelfSupervisedAlignment,
author = {Rhydian Windsor and
Amir Jamaludin and
Timor Kadir and
Andrew Zisserman},
title = {Self-Supervised Multi-Modal Alignment for Whole Body Medical Imaging},
booktitle = {MICCAI},
year = {2021}
}
The preprocessing occurs in 3 stages:
- Download the raw DXA and MRI DICOMS from the UK Biobank servers.
- Extract and process the files (and stitch the MRI scan parts together).
- Extract the mid-coronal slices from the MRI scans to reduce storage.
The raw data for this is extracted from the UK Biobank. We use whole body DXA images (field ID 20158) & Dixon technique MRI (field ID 20201). Please note that accessing this data requires an application to the Biobank (see here for more information).
Both these fields can be bulk downloaded using the ukbfetch script, made available by the Biobank.
Bash scripts to perform this is given in download_bb_dxas.py and download_bb_mris.py.
Please ensure a valid UK biobank authentication key (usually named .ukbkey) is available in this directory to perform the download. The given scripts are not parallelized, and will take around 3 days to complete, although upto 10 parallel downloads are allowed by the Biobank, so this could be reduced to around 8 hours by running across several threads.
The scripts can be run (in this case using /work/rhydian/ as a data directory) via
python download_bb_dxas.py -a /work/rhydian/UKBiobank/.ukbkey -o /work/rhydian/UKBB_Downloads/dxas/ -e /work/rhydian/UKBiobank/ukbb_eids.txt
and
python download_bb_mris.py -a /work/rhydian/UKBiobank/.ukbkey -o /work/rhydian/UKBB_Downloads/mris/ -e /work/rhydian/UKBiobank/ukbb_eids.txt
Now the raw scans have been downloaded the next step is to unzip them and stitch the axial slices of the MRI scans together.
To unzip and save the DXA files as pickle objects, run
python unzip_dxas.py -i /work/rhydian/UKBB_Downloads/dxas/ -o /work/rhydian/UKBB_Downloads/dxas-processed/
To unzip the MRI DICOMS, stitch them together and save them as a pickle object, run
python unzip_mris.py -i /work/rhydian/UKBB_Downloads/mris/ -o /work/rhydian/UKBB_Downloads/mris-processed/
Again, this is fairly slow but can be easily parallelized depending on compute resources available. An
example SBATCH script for performing this on slurm systems is given in unzip_mris_slurm.sh.
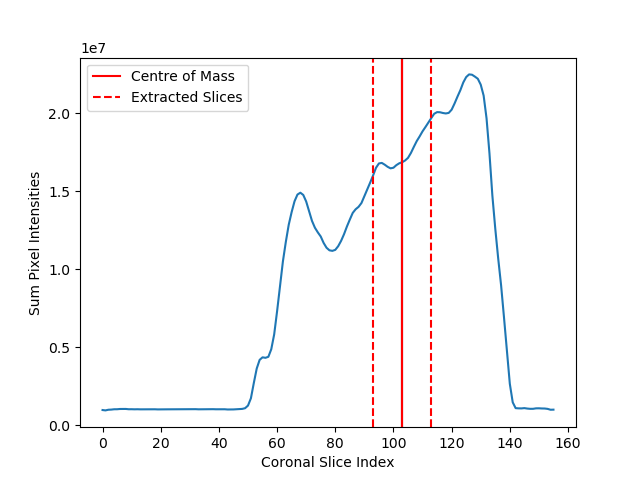
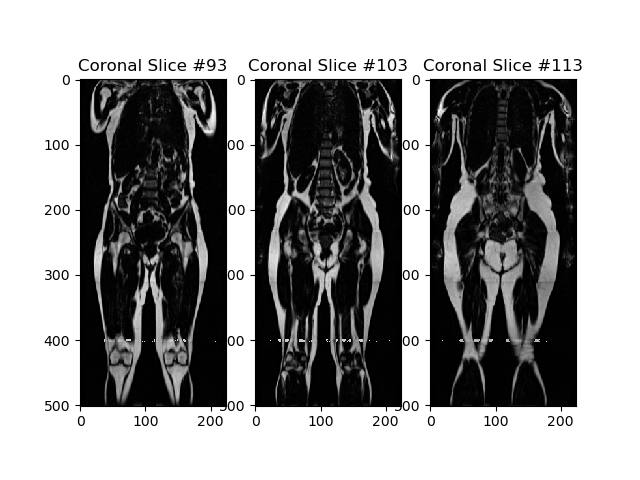
Finally, to reduce GPU memory constraints, the mid sagittal slices only are extracted from the MR scans. In the paper this is done by first detecting the spine in the scans using SpineNetV2 and using this detection to synthesize a mid spine coronal slice. However, since SpineNetV2 is not currently publically available, we will find the centre of the scan by finding the centre of mass of the coronal slices intensity histogram in the fat scan. For training-time augmentation, we also extract 5 neighbouring slices on each side (with 2 slices of spacing between each):
This is done by executing the following command
python extract_mri_mid_cor_slices.py -i /work/rhydian/UKBB_Downloads/mris-processed/ -o /work/rhydian/UKBB_Downloads/mri-mid-corr-slices/
At this point, mris-processed,mris and dxas can be deleted to save storage space.