Pytorch implementation for scPretrain: Multi-task self-supervised learning for cell type classification.
This code has been written using PyTorch >= 1.5.1. If you use any source codes or datasets included in this toolkit in your work, please cite the following paper. The bibtex is listed below:
@article{scPretrain,
title={scPretrain: Multi-task self-supervised learning for cell type classification},
author={Zhang, Ruiyi and Luo, Yunan and Ma, Jianzhu and Zhang, Ming and Wang, Sheng},
journal={arXiv preprint arXiv:2011.xxxxx},
year={2020}
}
Rapidly generated scRNA-seq datasets enable us to understand cellular differences and the function of each individual cell at single-cell resolution. Cell type classification, which aims at characterizing and labeling groups of cells according to their gene expression, is one of the most important steps for single-cell analysis. To facilitate the manual curation process, supervised learning methods have been used to automatically classify cells. Most of the existing supervised learning approaches only utilize annotated cells in the training step while ignoring the more abundant unannotated cells. In this paper, we proposed scPretrain, a multi-task self-supervised learning approach that jointly considers annotated and unannotated cells for cell type classification. scPretrain consists of a pre-training step and a fine-tuning step. In the pre-training step, scPretrain uses a multi-task learning framework to train a feature extraction encoder based on each dataset’s pseudo-labels, where only unannotated cells are used. In the fine-tuning step, scPretrain fine-tunes this feature extraction encoder using the limited annotated cells in a new dataset. We evaluated scPretrain on 60 diverse datasets from different technologies, species and organs, and obtained a significant improvement on both cell type classification and cell clustering. Moreover, the representations obtained by scPretrain in the pre-training step also enhanced the performance of conventional classifiers such as random forest, logistic regression and support vector machines. scPretrain is able to effectively utilize the massive amount of unlabelled data and be applied to annotating increasingly generated scRNA-seq datasets.
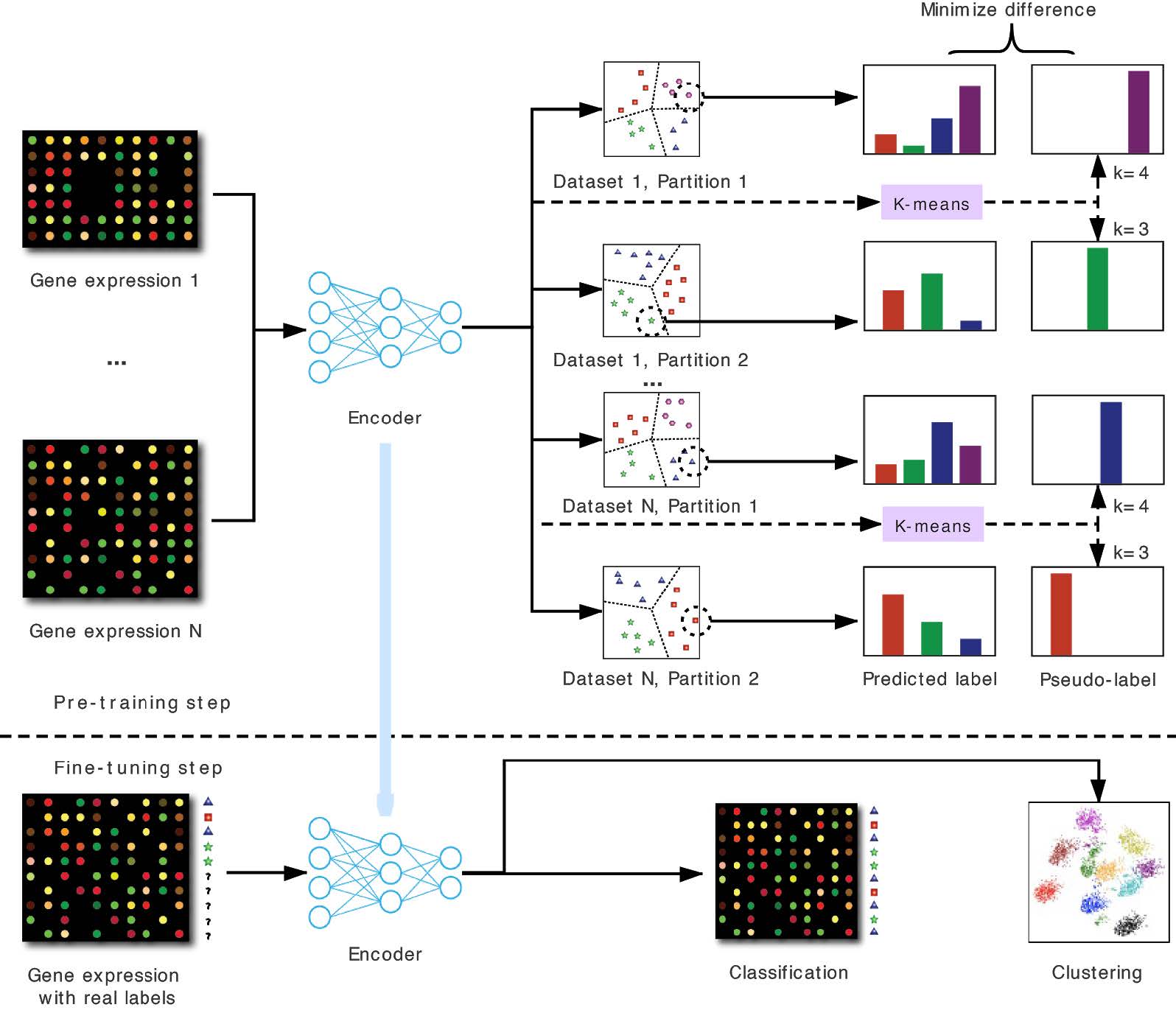
In the pre-training step, scPretrain assigns a pseudo-label to each cell using K-means. These pseudo-labels are used to train a feature extraction encoder, which is shared by different datasets and different partitions in a multi-task learning framework. In the fine-tuning step, this encoder is used to embed cells in a new dataset to low-dimensional representations, which are further used in downstream tasks such as cell clustering and cell type classification.
pip install -r requirements.txt
Automatically download from https://cblast.gao-lab.org/download. Inside folder: /dataset/
Pre-training step
Phase 1: pre-train on pseudo-labels generated by clustering on input. Output model name required by --save_model.
python main.py --cuda --is_pretrain --kmeans Phase 2: pre-train on pseudo-labels generated by clustering on output. Pre-trained model name required by --pretrained_model.
python main.py --cuda --is_pretrain --kmeans --pretrain_outputWe have already pre-trained a model and it can be downloaded from the following link:
https://drive.google.com/file/d/1OjuxWMm9F-ElA_SQx1kRg00XcSWoI6yQ/view?usp=sharing
After downloading the model above, it can be used for the following fine-tune step directly.
Fine-tuning step: automatically test on 60 datasets. Pre-trained model name required by --pretrained_model.
python main.py --cuda --kmeans Test other off-the-shelf classifiers (e.g SVM). Classifier name required by --clf.
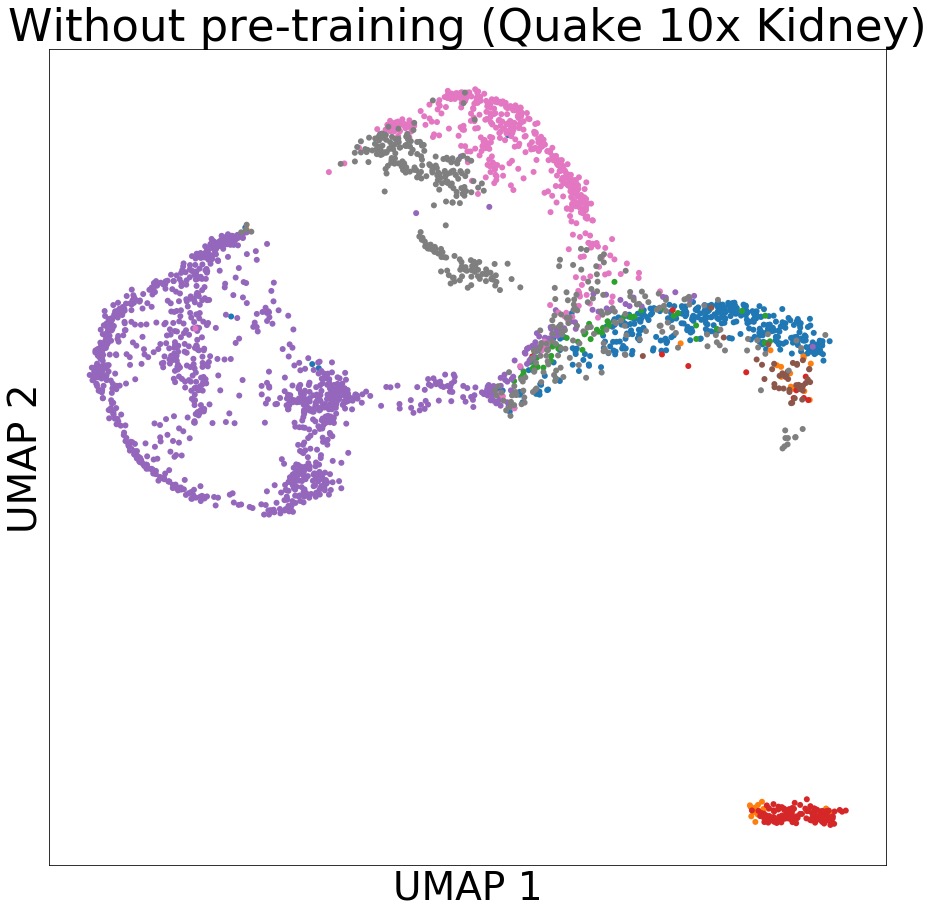
python rf_svm_lr.py --clf svm Produce cluster results using scPretrain generated representations and UMAP reducer.
python clus.py Create the scatter plots of Accuracy, Cohen's Kappa, AUPRC, AUROC, clustering etc. (Remember to do clustering and produce off-the-shelf classifier results first in order to create related figures)
>>> python fig.pySample figure:
To automatically use our code on other datasets, the dataset d should be a .h5 file and have following attributes:
d['exprs']['data']: gene expression data list.
d['exprs']['indices']: column indices for each data in d['exprs']['data'].
d['exprs']['indptr']: positions when row indices change in d['exprs']['indices'].
d['exprs']['shape']: the shape of the data matrix.
d['var_names']: gene names of the data matrix.
d['obs']['cell_ontology_class']: cell type for each gene expression vector.
Then put the d.h5 file in /dataset/, and add d in data.dataset_name. Then follow the fine-tuning step instruction above. (Note that we only support human and mouse gene datasets now).
To use numpy array finetune datasets, set the np_input argument to the path of a folder that includes 3 files: feature.npy, label.npy, gene_list.npy.
To use the anndata finetune datasets, set the ann_input argument to the path of the h5ad file.
For questions about the data and code, please contact zhangruiyi@pku.edu.cn. We will do our best to provide support and address any issues. We appreciate your feedback!