The following link is a link to the GitHub repository of the self assigned final project in the course Visual Analytics (F23.147201U023.A). Within the GitHub repository all necessary code are provided to reproduce the results of the project.
https://github.com/rikkeuldbaek/SkinCancerClassifier_VA_FP
For this final project I have worked with a collection of dermatoscopic images with different categories of pigmented skin lesions, i.e., skin cancer, in order to build a classifier that is able to segment and classify different diagnostic categories of skin cancer. The nature of the data is quite unbalanced, hence two identical classifiers have been designed to classify a balanced dataset of skin cancer and an unbalanced dataset of skin cancer. The performance of the two identical classifiers is evaluated in order to review the impact of varying data distributions. This repository contains source code which train two identical pretrained CNN on the skin cancer dataset (the HAM10000 dataset), classifies the diagnostic categories of skin cancer and produce a classification reports and a training/validation history plots.
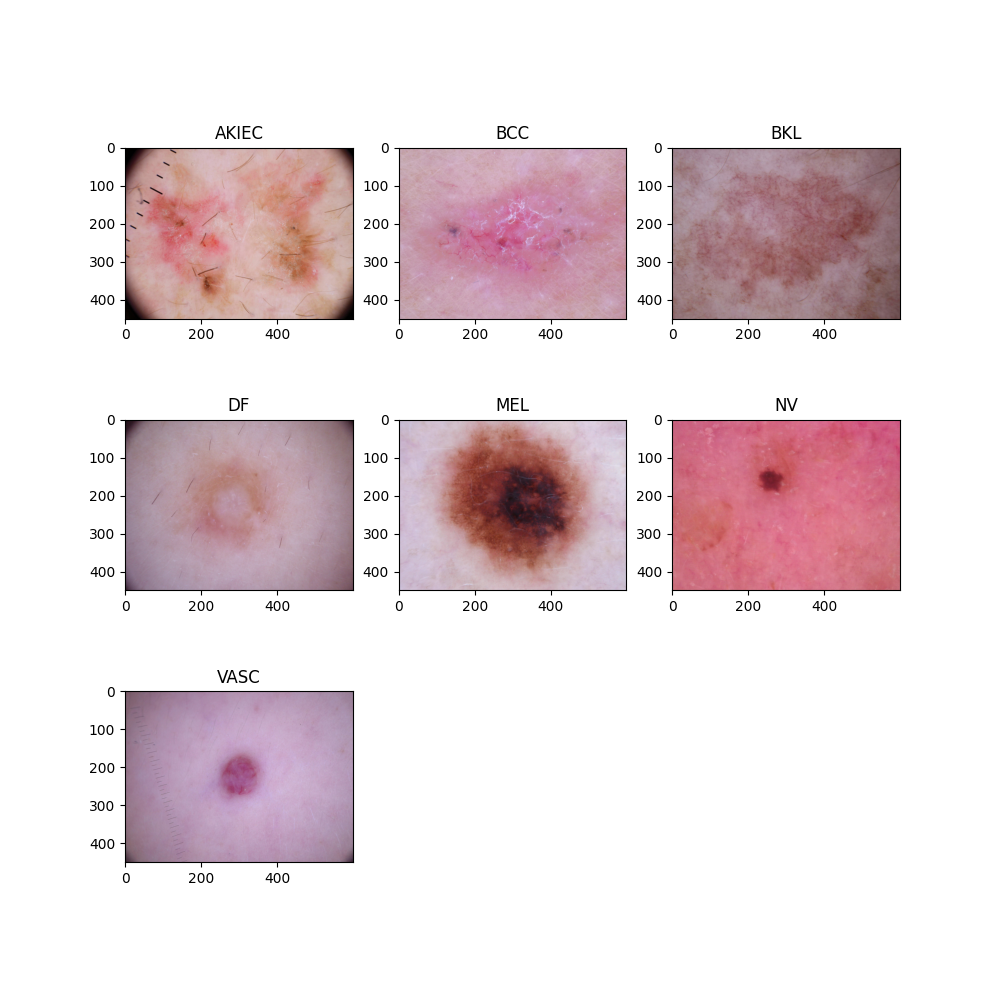
For this final project I have used the HAM10000 dataset ("Human Against Machine with 10000 training images"). This dataset contains 10015 multi-source dermatoscopic images of common pigmented skin lesions, also known as skin cancer. This collection of images of skin cancer includes seven representative diagnostic categories within the domain of pigmented lessions. These seven categories are: Actinic keratoses and intraepithelial carcinoma / Bowen's disease (AKIEC), basal cell carcinoma (BCC), benign keratosis-like lesions (solar lentigines / seborrheic keratoses and lichen-planus like keratoses, BKL), dermatofibroma (df), melanoma (MEL), melanocytic nevi (NV) and vascular lesions (angiomas, angiokeratomas, pyogenic granulomas and hemorrhage, VASC) (Tschandl, 2018). Furthermore, the HAM10000 dataset consists of a Ground Truth .csv file matching each image filename to its diagnostic category. The plot below illustrates each of the seven diagnostic categories of skin cancer.
Plot 1: Diagnostic Categories of Skin Cancer
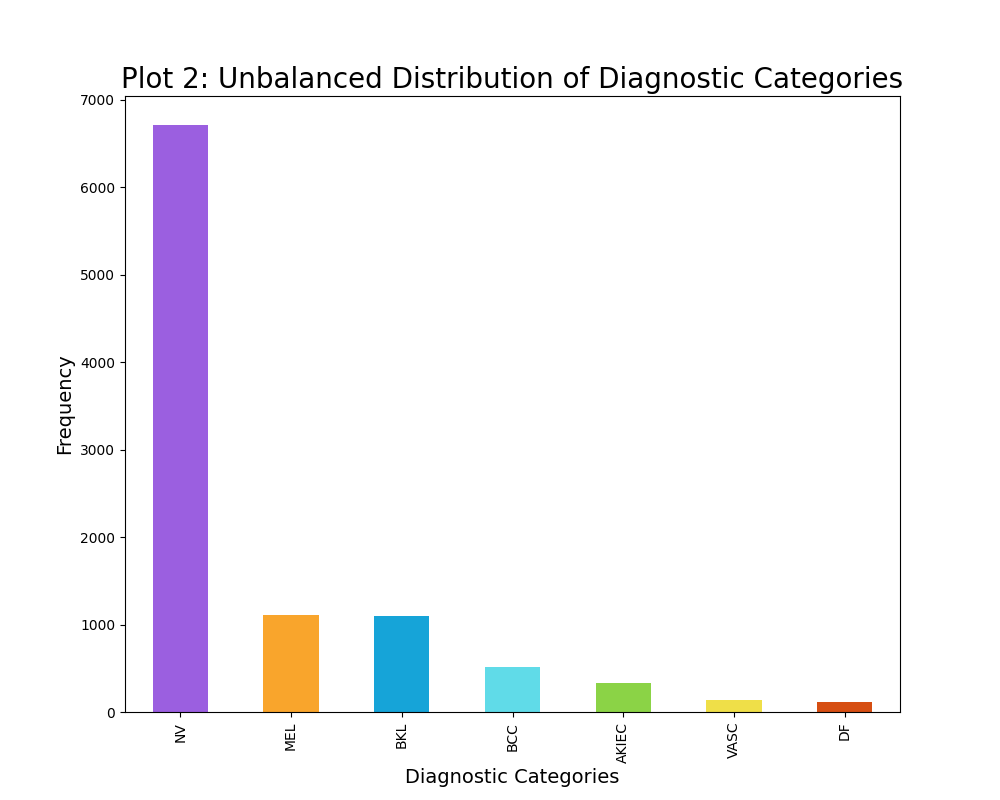
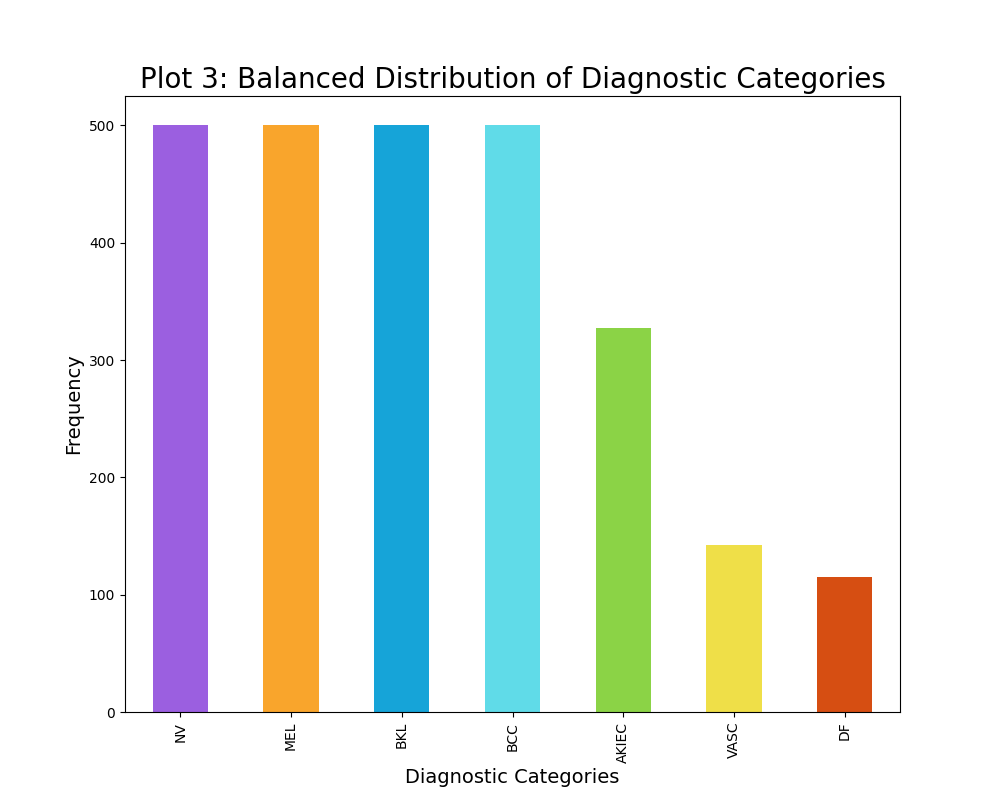
The distribution of the diagnostic categories of skin cancer was intitially rather unbalanced, as seen from plot 2. For instance the unbalanced data contained 6705 images of the diagnostic category NV, while containing 115 images of the diagnostic category DF. Thus, a maximum limit of 500 samples per diagnostic category was established, although some categories had less than 500 data points. This resulted in a slightly more balanced dataset, however it is not completely balanced out as visually evident from plot 3. Furthermore, this maximum limit of 500 samples per diagnostic category decreased the sample size substantially from 10015 images to 2584 images. This decrease in sample size have a great impact on the modelling, thus I have created two classifiers: one of which trains on the unbalaned data (10015 data points) and one of which trains on the balanced data (2584 data points). The varying results will be discussed in the result section.
 |
 |
The scripts of this project require a certain folder structure, thus the table below presents the required folders and their description and content.
| Folder name | Description | Content |
|---|---|---|
data |
images of skin cancer and .csv file with labels | archive/images/all_images,archive/GroundTruth.csv |
src |
model, data, and plot scripts | data.py, classifier_balanced.py, classifier_unbalanced.py, plot.py, unzip_data.py |
out |
classification reports and training/validation history plot | classification_report_balanced.txt,classification_report_unbalanced.txt, train_val_history_plot_balanced.png, train_val_history_plot_unbalanced.png |
readme_pngs |
plots for the readme file | balanced_distribution.png, unbalanced_distribution.png, diagnostic_categories.png |
utils |
helper functions | helper_func.py |
The unzip_data.py script located in src unzips the data into the datafolder. The data.py script located in src preprocesses the data and produces training, test, and validation data for both balanced and unbalanced data. The classifier_balanced.py and classifier_unbalanced.py scripts located in src produce skin cancer classifier models trained on balanced and unbalanced data, furthermore they produce classification reports and a training/validation history plots which are saved in the folder out. Helper functions for plotting are found in the folder utils and relevant plots for illustrations are found in the folder readme_pngs.
In order for the user to be able to run the code, please make sure to have bash and python 3 installed on the used device. The code has been written and tested with Python 3.9.2 on a Linux operating system. In order to run the provided code for this assignment, please follow the instructions below.
1) Clone the repository Please execute the following command in the terminal to clone this repository.
git clone https://github.com/rikkeuldbaek/SkinCancerClassifier_VA_FP.git2) Setup
Please execute the following command in the terminal to setup a virtual environment (VA_fp_env) and install packages.
bash setup.sh3) Download the data and unzip the file
Please download the HAM10000 dataset from Kaggle (HAM10000 dataset
), and store the archive.zip zipfile in the data folder in this repository. Please run the following script in order to unzip file into the data folder. The command must be executed in the terminal. This may take a few minutes since the files in the zipfile are around 3GB in total.
python src/unzip_data.py Please execute the following command in the terminal to automatically run the data.py, classifier_balanced.py, andclassifier_unbalanced.py scripts.
bash run.shThe skin cancer classifier have the following default arguments stated in the table below. These arguments can be modified and adjusted in the run.sh script. If no modifications are added, default parameters are run. In case help is needed, please write --help in continuation of the code below instead of writing an argument.
python src/classifier.py #add arguments here or --helpBoth classifier_balanced.py and classifier_unbalanced.py take the following arguments:
| Argument | Type | Default |
|---|---|---|
| --target_size | int | (224,224) |
| --horizontal_flip | bool | True |
| --vertical_flip | bool | True |
| --zca_whitening | bool | True |
| --shear_range | float | 0.2 |
| --zoom_range | float | 0.2 |
| --rotation_range | int | 20 |
| --rescale_1 | float | 1. |
| --rescale_2 | float | 225. |
| --batch_size | int | 30 |
| --n_epochs | int | 30 |
| --class_mode | str | categorical |
| --pooling | str | avg |
| --input_shape | int | (224,224,3) |
| --monitor | str | val_loss |
| --patience | int | 5 |
| --restore_best_weights | bool | True |
| --nodes_layer_1 | int | 300 |
| --nodes_layer_2 | int | 150 |
| --activation_hidden_layer | str | relu |
| --activation_output_layer | str | softmax |
| --initial_learning_rate | float | 0.01 |
| --decay_steps | int | 10000 |
| --decay_rate | float | 0.9 |
| --loss | str | categorical_crossentropy |
The data.py takes the following arguments:
| Argument | Type | Default |
|---|---|---|
| --train_split_bal | float | .70 |
| --val_split_bal | float | .15 |
| --train_split_ubal | float | .70 |
| --val_split_ubal | float | .15 |
The target_size and the input_shape argument must be specified without commas in the run.sh script, please see following command for an example of such:
python src/classifier.py --target_size 224 224 --input_shape 224 224 3Similarly it is very important to note that the data.py is automatically called upon when running both classifier scripts, thus the arguments for data.py must be parsed to the classifier_balanced.py and classifier_unbalanced.py script in run.sh:
python src/classifier_balanced.py --classifier_arguments --data_arguments
python src/classifier_unbalanced.py --classifier_arguments --data_argumentsClassification report (open link to see)
The model shows an accuracy of 49%. Furthermore, the model seems to be best at classifying NV, MEL, and BCC with F1 scores of 63%, 55%, 51% respectively. This makes sense, as the model sees more images of these three diagnostic categories, and thus the model become more exposed to learning them. The model seems to be worst at classifying DF, BKL, AKIEC, VASC with F1 scores of 0%, 30%, 45% and 48% respectively. This also makes sense, as the model is exposed to fewer images of these diagnostic categories. However it is not the case of BKL. BKL actually have almost as many images in the test data as the best predicted diagnostic categories, but it only gains an F1 score of 30%.
The reason for this may be that the features of BKL are not that salient and may resemble other types of skin cancer, and thus becomes harder to classify (see plot 1 of diagnostic categories). It is also evident from the precision and recall score that the skin cancer classifier has a high number of false positives and false negatives, which may be caused by a yet not totally balanced dataset.
Training and validation history plot
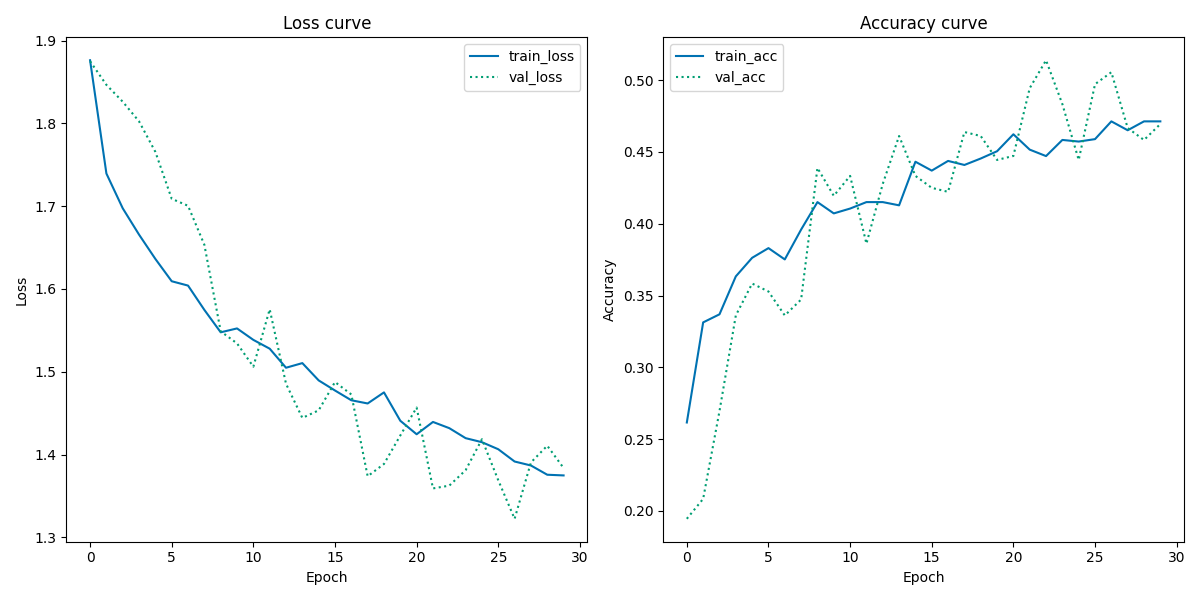
From the Loss Curve plot, the validation loss looks very noisy while the training loss looks decent, indicating a unrepresentative validation dataset. Furthermore, the validation loss curve is mainly below the training loss curve towards the end, suggesting that the model finds it easier to predict the validation dataset than the training dataset.
From the Accuracy Curve plot, similar noisy tendencies in the validation accuracy are present. However, the training accuracy curve seems to have reached a point of stability where no more new knowledge is acquired. Overall, the performance of the model is not ideal, as it looks like it performs better on unseen data than seen data, which makes very little sense.
Classification report (open link to see)
The model shows an accuracy of 73%. Furthermore, the model seems to be best at classifying NV, VASC, MEL, and BCC with F1 scores of 88%, 47%, 44%, and 43% respectively. However, the model is not necessarily exposed to a lot of images within the two diagnostic categories VASC and BCC. Moreover, it must be noticed that three out of four of the best F1 scores are below chance level.
The model seems to be worst at classifying DF, AKIEC, and BKL with F1 scores of 0%, 26%, 33% respectively. It makes sense, that the model performs bad on DF and AKIEC with relatively few images in the test set, and these two diagnostic categories have very sublte features that could resemble other types of skin cancer. However BKL has a descent amount of images in the test set, but the model seems to find it hard to predict the diagnostic category accurately. The reason for this may be the same as mentioned in the above balanced classification report section.
Training and validation history plot
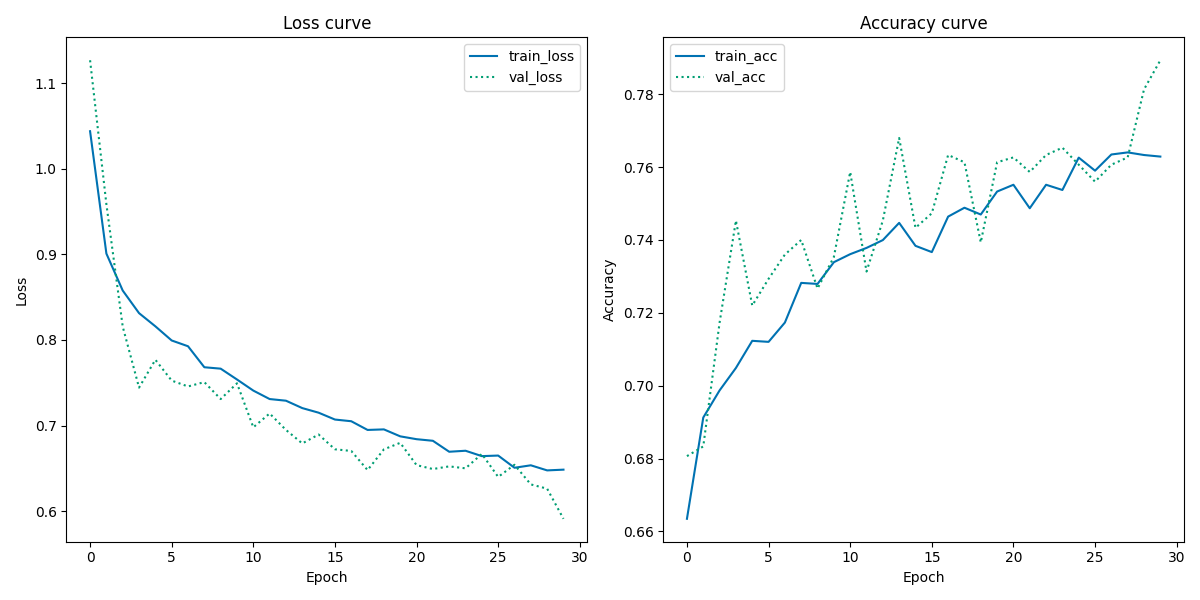
From the Loss Curve plot, both the validation loss and training loss show similar gradually descending tendencies in their slope with only a little gap between them. This may indicate that there no or little overfitting. There seems to be a little discrepancy between the training loss and validation loss towards the end since the slope of the training loss appears to be reaching a point of stability around 28-29 epochs, indicating that the model is not capable of further learning, while the validation loss seems to be decreasing further. Also the validation loss looks some what noisy, suggesting a unrepresentative validation dataset.
From the Accuracy Curve plot, there are similar tendencies of discrepancy towards the final epochs: the training accuracy seems to have reached a point of stability (no more learning is acquired), while the validation accuracy increase further. Like the Loss Curve plot, the validation accuracy is still very noisy, indicating unrepresentative validation dataset. Moreover, most of the validation accuracy curve is above the training accuracy curve, making it appear as if the model performs better on unseen data than seen data.
In summary, the overall accuracy performance of the skin cancer classifier is best using unbalanced data, however there are some pitfalls to it as mentioned above. The learning curves of the skin cancer classifier show same poor performance, which overall indicate that the validation dataset must be reconsidered in the data preprocessing steps in the workflow.
Tschandl, P. (2018). The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions (ViDIR Group, Ed.; V4 ed.). Harvard Dataverse. https://doi.org/10.7910/DVN/DBW86T