| Branch |  |
 |
|---|---|---|
master |
||
develop |
R package to work with PureseqTM.
Because PureseqTM only works on Linux, pureseqtmr only works on Linux.
library(pureseqtmr)
# Use an example proteome
fasta_filename <- system.file(
"extdata",
"UP000464024.fasta",
package = "pureseqtmr"
)
# Predict the topology
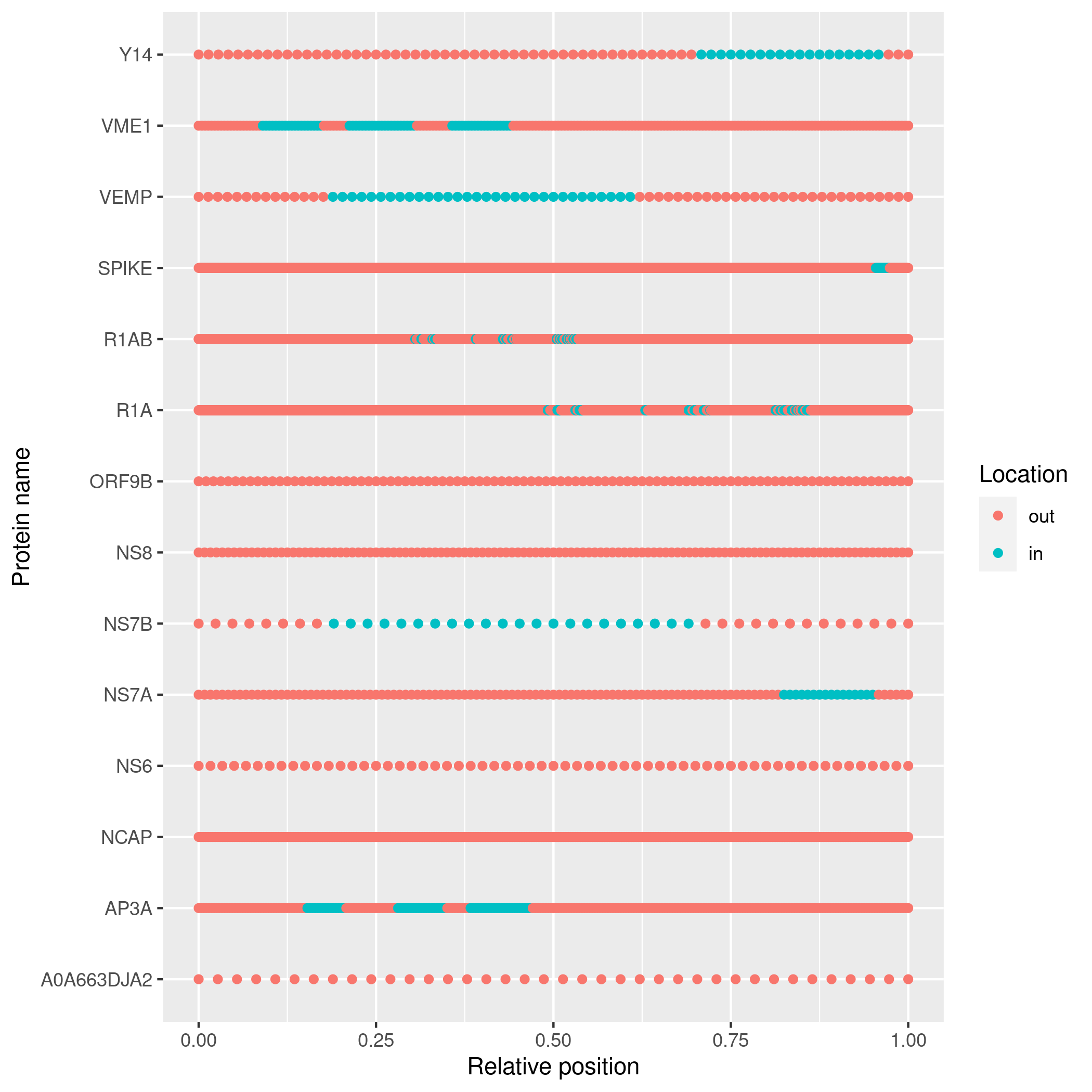
topology <- predict_topology(fasta_filename)
# Simplify the protein names
topology$name <- stringr::str_match(
string = topology$name,
pattern = "..\\|.*\\|(.*)_SARS2"
)[,2]
# Plot the topology
plot_topology(topology)Due to CRAN policy, pureseqtmr cannot install PureseqTM.
As a workaround, the non-CRAN
pureseqtmrinstall
can be used.
To install PureseqTM:
remotes::install_github("richelbilderbeek/pureseqtmrinstall")
pureseqtmrinstall::install_pureseqtm()Combine predict_topology and save_tibble_as_fasta_file:
library(pureseqtmr)
save_tibble_as_fasta_file(
predict_topology(fasta_filename = "my.fasta"),
fasta_filename = "my.topo"
)
These are the ones I use:
- tmhmm:
predict membrane protein topology at three levels (inside, outside, membrane),
do need registration form, as it is for academic users only. See here for a run-time speed comparison. Spoiler: PureseqTM is approximately 20x slower than TMHMM
pureseqtmr gives an error.
PureseqTM ignores the selenocystein, as it does with any character.
pureseqtmr package gives an error.
PureseqTM prints out a message and returns a zero-length topology.
See CONTRIBUTING, at Submitting use cases
See CONTRIBUTING, at 'Submitting code'
See CONTRIBUTING, at 'Submitting bugs'
Sure, just add an Issue. Or send an email.
Article about PureseqTM:
- Wang, Qing, et al. "Efficient and accurate prediction of transmembrane topology from amino acid sequence only." bioRxiv (2019): 627307.