This software, developed at the REDS institute in the context of the MOVABLE project (MicrOscopic VisuAlization of BLood cElls for the Detection of Malaria and CD4+), is aimed at detecting Plasmodium falciparum parasites in microscope images of blood samples and computing the corresponding parasitemia.
We actively seek the contribution of the community, do not hesitate to contact us if you are willing to help. Also, if you find a bug or have an idea for an interesting feature, please open a ticket!
For further information on the project, please refer to the project's webpage.
The software is composed by two parts:
- the classifier, which learns how to detect parasites on a set of manually-classified images and then applies these criteria on unseen images;
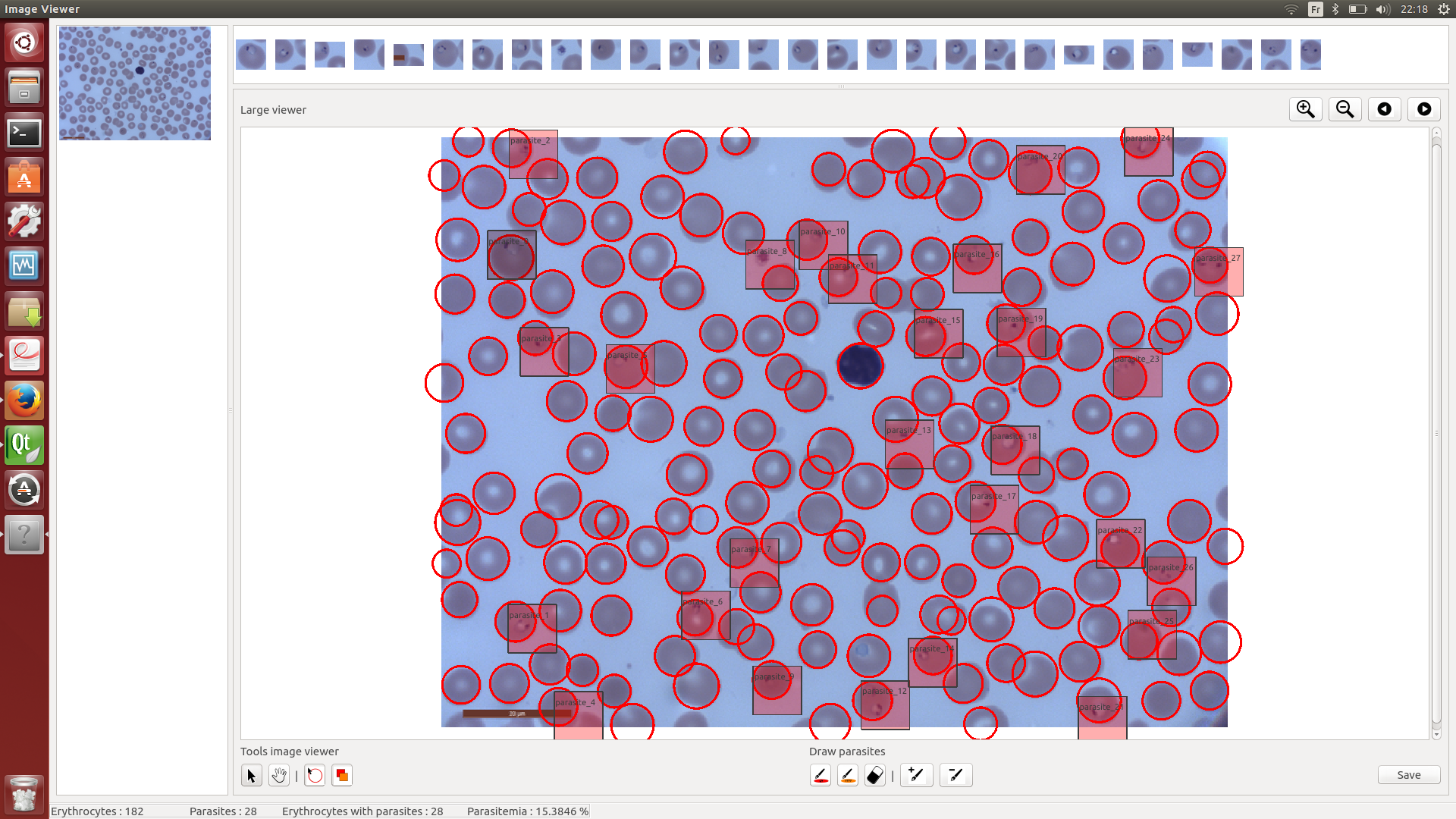
- the GUI, which takes as input an already-classified image and allows to visualize the parasites and fix misdetections.
The learning process is based on the KernelBoost algorithm, an intuitive description of which is given in this document. Part of the code has been adapted from the SQBlib library.
The visual interface allows the user to manually fix automated segmentations and then export these modifications to a file, which can then be fed back to the classifier for a retraining.
To compile the classifier, please follow the instructions in the INSTALL file.
The GUI is a QT project and needs the qmake compiler. The documentation to install the QT framework and its compiler is available on the official QT web site. The configuration of the project is stored in the file movable_ui.pro.
To prepare the data, create a directory (whose path has to be specified in the configuration files) and put in it 5 files:
- a list of paths for training images (absolutes or relatives to the current execution directory)
- a list of paths for training masks (absolutes or relatives to the current execution directory)
- a list of paths for training ground-truths (absolutes or relatives to the current execution directory)
- a list of paths for test images (absolutes or relatives to the current execution directory)
- a list of paths for test masks (absolutes or relatives to the current execution directory) The filenames are free (you have, however, to specify them in the configuration files), and the name of the images too, but the order of the paths in the files is not: the i-th image has to match with the i-th mask and the i-th ground-truth --- that is, all the three files for a given image have to be in the same row number on the three different list files. They also have to match in size.
The paths of the datasets MUST be specified in the configuration files (train_config.json and test_config.json)
To execute the software, enter the directory build/src/train and execute ./train_movable Training options can be altered in the train_config.json file
Once the classifier has been trained, it can be used to test on images by entering build/src/test and executing ./test_movable
The classifier outputs black&white images where white areas mark detected parasites. These segmentation can be visualized and modified via the the GUI available in movable/gui.
GUI usage video
MOVABLE is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
MOVABLE is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.