Probabilistic Annotation INtegraTOR
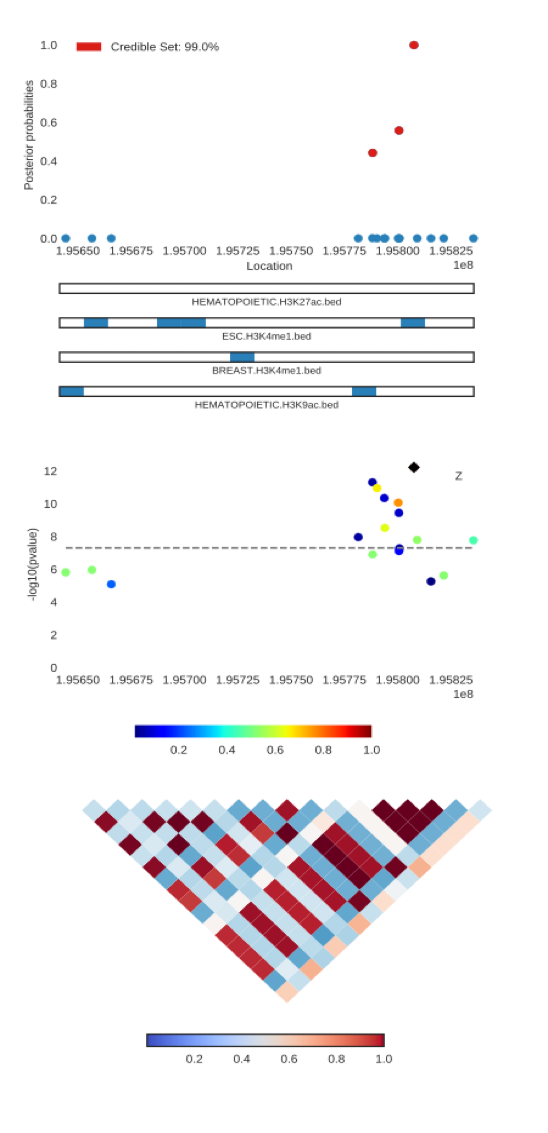
Announcing PAINTOR v3.0! The new version has enhancements that improve computational effiency, statistical robustness, as well as having expanded functionality to leverage multiple traits. In adddition, we have developed a visualiziation tool, PAINTOR-CANVIS, to produce publication-ready plots for the output of PAINTOR as seen below.
For legacy purposes, we leave available PAINTOR 2.1, though we recommend using this latest version for most accurate results.
We provide a command line implementation of the PAINTOR frameworks described in Kichaev et al. (PLOS Genetics, 2014), (American Journal of Human Genetics, 2015), and (Bioinformatics, 2016). Briefly, PAINTOR is a statistical fine-mapping method that integrates functional genomic data with association strength from potentially multiple populations (or traits) to prioritize variants for follow-up analysis. The software runs on multiple fine-mapping loci and/or populations/traits simultaneously and takes as input the following data for each set of SNPs at a locus
- Summary Association Statistics (Z-scores)
- Linkage Disequilibrium Matrix/Matrices (Pairwise Pearson correlations coefficients between each SNP)
- Functional Annotation Matrix (Binary indicator of annotation membership (i.e. if entry {i,k} = 1, then SNP i is a member of annotation K).
- Outputs a probability for a SNP to be causal which can subsequently be used to prioritize variants
- Can model multiple causal variants at any risk locus
- Leverage functional genomic data as a prior probability to improve prioritization
- This prior probability is not pre-specified, but rather, learned directly from the data via Empirical Bayes.
- Quantify enrichment of causal variants within functional classes
- Enables users to unbiasedly select from a (potentially) large pool functional annotations that are most phenotypically relevant
- Fully Bayesian treatment of causal effect sizes
- (optional) Model population-specific LD patterns when doing multi-ethnic fine-mapping.
- (optional) Joint inference across traits when doing multi-trait fine-mapping.
- (optional) Approximate inference via Importance Sampling.
For detailed information about input file formats, command line flags, and recommended analysis pipelines please see the wiki
The software has two dependencies: [1] Eigen v3.2 (matrix library) [2] NLopt v2.4.2 (optimization library) which are packaged with PAINTOR in order to simplify installation. Please see the Eigen homepage and NLopt homepage for more information. Note that compiling requires gcc V4.9 (or greater).
For quick installation:
git clone https://github.com/gkichaev/PAINTOR_V3.0.git
cd PAINTOR_V3.0
bash install.sh
This will create an executable "PAINTOR". Sample data is provided with the package. To test that the installation worked properly, type:
./PAINTOR -input SampleData/input.files -in SampleData/ -out SampleData/ -Zhead Zscore -LDname ld -enumerate 2 -annotations DHS
If everything worked correctly the final sum of log Bayes Factors should be: 654.233501
For quick start simply type:
./PAINTOR
We have compiled library of functional annotations that you may find useful. This large compendium includes .bed files for most of the Roadmap/ENCODE data as well as other regulatory and genic annotations. Please see the [wiki] (https://github.com/gkichaev/PAINTOR_V3.0/wiki/2b.-Overlapping-annotations) for more information and download link.