DeCiFer
DeCiFer is an algorithm that simultaneously selects mutation multiplicities and clusters SNVs by their corresponding descendant cell fractions (DCF), a statistic that quantifies the proportion of cells which acquired the SNV or whose ancestors acquired the SNV. DCF is related to the commonly used cancer cell fraction (CCF) but further accounts for SNVs which are lost due to deleterious somatic copy-number aberrations (CNAs), identifying clusters of SNVs which occur in the same phylogenetic branch of tumour evolution.
The full description of the algorithm and its application on published cancer datasets are described in
Gryte Satas†, Simone Zaccaria†, Mohammed El-Kebir†,* and Ben Raphael*, 2021
† Joint First Authors
* Corresponding Authors
The results of the related paper are available at:
This repository includes detailed instructions for installation and requirements, demos and tutorials of DeCiFer, a list of current issues, and contacts. This repository is currently in a preliminary release and improved versions are released frequently. During this stage, please keep checking for updates.
Contents
Algorithm
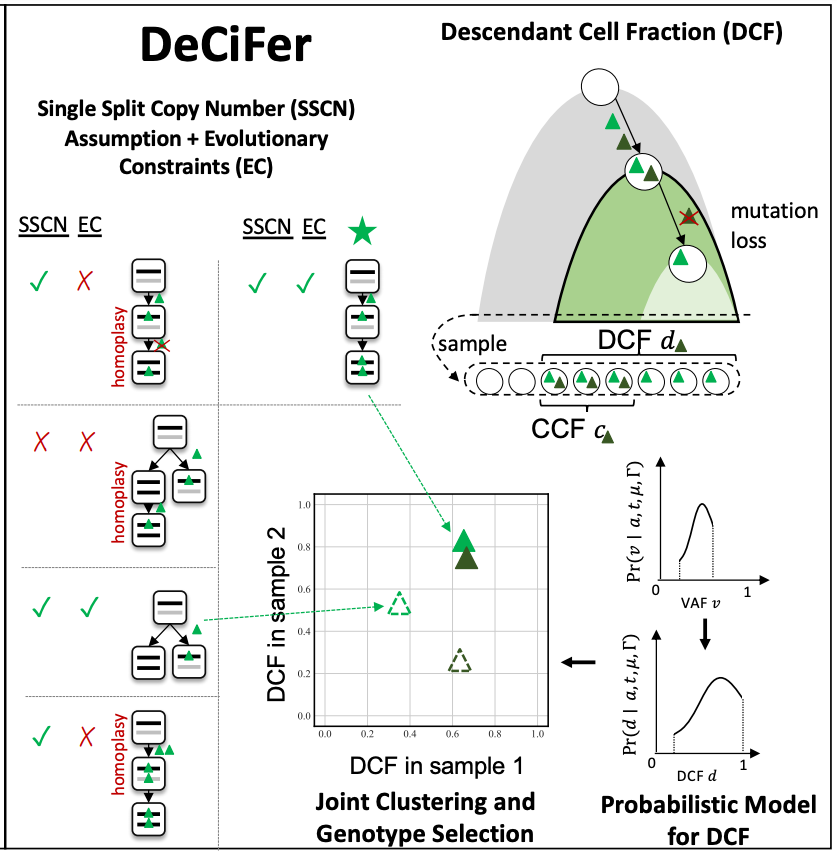
DeCiFer uses the Single Split Copy Number (SSCN) assumption and evolutionary constraints to enumerate potential genotype sets. This allows DeCiFer to exclude genotype sets with constant mutation multiplicity (CMM) that are not biologically likely (red crosses) and include additional genotype sets (green star) that are. DeCiFer simultaneously selects a genotype set for each SNV and clusters all SNVs based on a probabilistic model of DCFs, which summarize both the prevalence of the SNV and its evolutionary history.
Installation
DeCiFer is mostly written in Python 2.7 and has an optional component in C++. The recommended installation is through conda but we also provide custom instructions to install DeCiFer in any Python environment.
Automatic installation
The recommended installation is through bioconda and requires conda, which can be easily and locally obtained by installing one of the two most common freely available distributions: anaconda or miniconda. Please make sure to have executed the required channel setup for bioconda. Thus, the following one-time one-line command is sufficient to fully install DeCiFer within a virtual conda environment called decifer:
conda create -n decifer decifer -yAfter such one-time installation, DeCiFer can be executed in every new session after activating the decifer environment as follows:
conda activate deciferManual installation
DeCiFer can also be installed in a conda environment directly from this repo. Thus, the following one-time commands are sufficient to fully install DeCiFer within a virtual conda environment called decifer from this Git repo:
git clone https://github.com/raphael-group/decifer.git && cd decifer/
conda create -c anaconda -c conda-forge -n decifer python=2.7 numpy scipy matplotlib-base pandas seaborn -y
pip install .Custom installation
DeCiFer can be installed with pip by the command pip install . in any Python2.7 environment with the following packages or compatible versions:
| Package | Tested version | Comments |
|---|---|---|
| numpy | 1.16.1 | Efficient scientific computations |
| scipy | 1.2.1 | Efficient mathematical functions and methods |
| pandas | 0.20.1 | Dataframe management |
| matplotlib | 2.0.2 | Basic plotting utilities |
| seaborn | 0.7.1 | Advanced plotting utilities |
Installation of C++ component
DeCiFer includes C++ code to enumerate state/genotype trees. The dependencies for this code are as follows.
| Package | Tested version | Comments |
|---|---|---|
| cmake | >= 2.8 | Build environment |
| lemon | 1.3.1 | C++ graph library |
| boost | >= 1.69.0 | C++ library for scientific computing |
To build this code, enter the following commands from the root of the repository:
mkdir build
cd build
# OPTIONAL: specify lemon and/or Boost paths if not detected automatically.
cmake ../src/decifer/cpp/ -LIBLEMON_ROOT=/usr/local/ -DBOOST_ROOT=/scratch/software/boost_1_69_0/
makeUsage
DeCiFer can be executed using the command decifer, whose manual describes the available parameters and argument options. See more details below.
- Required input data
- Optional input data
- Output
- System requirements
- Demos
- Reccomendations and quality control
Required input data
DeCiFer requires two input data:
- Input mutations with nucleotide counts and related copy numbers in a tab-separated file (TSV) with three header lines ((1) The first specifies the number of mutations; (2) The second specifies the number of samples; and (3) The third is equal to:
#sample_index sample_label character_index character_label ref var) and where every other row has the following values for every mutation in every sample:
| Name | Description | Mandatory |
|---|---|---|
| Sample index | a unique number identifying the sample | Yes |
| Sample label | a unique name for the sample | Yes |
| Mutation index | a unique number identifying the mutation | Yes |
| Mutation label | a unique name identifying the mutation | Yes |
| REF | Number of reads with reference allele for the mutation | Yes |
| ALT | Number of reads with alternate allele for the mutation | Yes |
| Copy numbers and proportions | Tab-separated A B U where A,B are the inferred allele-specific copy numbers for the segment harboring the mutation and U is the corresponding proportion of cells (normal and tumour) with those copy numbers. Groups of cells/clones with the same allele-specific copy numbers must be combined into a single proportion. |
Yes |
| Additional copy numbers | An arbitrary number of fields with the same format as of Copy numbers and proportions describing the proportions of cells with different copy numbers. Note that all proportions should always sum up to 1. |
No |
- Input tumour purity in a two-column tab-separated file where every row
SAMPLE-INDEX TUMOUR-PURITYdefines the tumour purityTUMOUR-PURITYof a sample with indexSAMPLE-INDEX.
For generating the input files for DeCiFer, please see the scripts directory for more information. Examples may be found in the data directory.
Optional input data
DeCiFer can use the following additional and optional input data:
1. Data for fitting beta-binomial distributions to read count data
To use beta-binomial distributions to cluster mutations (default is binomial), pass the --betabinomial flag to decifer along with 2 additional arguments, --snpfile and --segfile, which are used to specify the locations of 2 files that contain information to parameterize the beta-binomial for each sample.
The file passed to DeCiFer via --snpfile contains information about the read counts of germline (not somatic) variants and has the following format:
| Field | Description |
|---|---|
SAMPLE |
Name of a sample |
CHR |
Name of the chromosome |
POS |
Genomic position in CHR |
REF_COUNT |
Number of reads harboring reference allele in POS |
ALT_COUNT |
Number of reads harboring alternate allele in POS |
The file passed to DeCiFer via --segfile, which specifies the allele-specific copy number per segment, is the same as the best.seg.ucn file used by the vcf_2_decifer.py python script that generates the input files for DeCiFer. Please simply specify the location of this file.
Custom state trees
Users may pass a file containing the set of all possible state trees for DeCiFer to evaluate. State trees have been pre-generated for the set of most common copy numbers, however a dataset might have a combination of copy numbers which has not been included. In this case, the user can use the command generatestatetrees to generate all the state trees needed for their dataset, for instance, following the instructions in the scripts directory. The script in this directory not only generates input files for decifer, but also a file called cn_states.txt that lists all the unique CN states for your data. This file may be used with generatestatetrees as shown in the scripts directory under the section "Adressing the "Skipping mutation warning"".
Output
DeCiFer's main output file (ending with _output.tsv) corresponds to a single TSV file encoding a dataframe where every row corresponds to an input mutation and with the following fields:
| Name | Description |
|---|---|
mut_index |
Unique identified for a mutation |
VAR_{SAMPLE} |
Variant sequencing read count of the mutation for every sample with index {SAMPLE} |
TOT_{SAMPLE} |
Total sequencing read count of the mutation for every sample with index {SAMPLE} |
VAR_{SAMPLE} |
Variant sequencing read count of the mutation for every sample with index {SAMPLE} |
cluster |
Unique identifier of the inferred mutation cluster |
state_tree |
Inferred state tree defined as a ->-separated edge list of genotypes |
cluster |
Unique identifier of the inferred mutation cluster; cluster 1 is the truncal cluster, and the next p clusters (where p is the number of samples) are sample-specific clusters, or SNVs that are unique to one of the p samples |
true_cluster_DCF{SAMPLE} |
Inferred true cluster DCF of the mutation in every sample with index {SAMPLE}; when execute in CCF-mode, DCF will be CCF instead; these values take the form cluster center;(lower cluster CI, upper cluster CI) |
point_estimate_DCF{SAMPLE} |
Point estimate of the mutation DCF in every sample with index {SAMPLE}; when execute in CCF-mode, DCF will be CCF instead |
cmm_CCF{SAMPLE} |
Inferred CCF of the mutation under the previous CMM assumption in every sample with index {SAMPLE} |
Explained |
;-separated list of all the clusters to which the mutation could be assigned |
LHs |
;-separated list of the negative-log likelihoods of assigned the mutation to all clusters in Explained |
For the column containing the true_cluster_DCF, the CIs correspond to the 95% credible interval of the posterior distribution of the DCF cluster center (Eqn 8 in manuscript and S23 in supplement) . These CIs have been corrected for multiple tests. Specifically, for each cluster, we find the lower CI by finding the X=[0.025/(number of hypothesis tests)] quantile, where the number of tests corresponds to (number of clusters)*(number of samples for patient). The same procedure is used for the upper CI, by finding the quantile that corresponds to 1-X.
These cluster CIs may also be found in the output file ending in _cluster.CIs.tsv. This file contains this information in a more condensed format, reporting only the upper and lower CIs for each cluster for each sample (in column f_lb and f_ub respectively). These numbers may contain "NaN" if no mutations were assigned to that particular cluster.
The file ending in _model_selection.tsv shows how decifer selected the best value of K clusters.
Lastly, the file ending in _Outliers_output.tsv contains SNVs that were flagged as outliers: the variant allele frequency (VAF) of the SNV was more than 1.5 (default) standard deviations away from the VAF of the assigned cluster center. Users may change this behavior via the --vafdevfilter option. This default behavior filters out noisy data or germline contamination that manifests as e.g. SNVs being assigned to the truncal cluster yet having very low DCF values in the point_estimate_DCF column of the output file.
System requirements
DeCiFer is highly parallelized in order to make efficient the extensive computations needed for clustering under a probabilistic model thousands of mutations across multiple tumour samples from the same patient. We recommend executing DeCiFer on multi-processing computing machines as the running time will scale down nearly proportionally with the number of parallel jobs, which can be specified with the argument -j. If the parameter is not specified, then DeCiFer will attempt to use all available CPUs; however, when using a computing cluster, we strongly recommend the user to always specifies -j in order to match the number of requested CPUs and avoid computing competition. Finally, note that also required memory also scales with the number of parallel processes; however in all previous tests on thousands of mutations with high number of parallel processes, DeCiFer never required more than 80GB of RAM. Please lower -j in case of exceeding memory.
Demos
Each demo is an exemplary and guided execution of a DeCiFer.Each demo is simultaneously a guided description of the entire example and a BASH script which can be directly executed to run the complete demo from this repository. As such, the user can both read the guided description as a web page and run the same script to execute the demo. At this time the following demos are available (more demos will be available soon):
| Demo | Description |
|---|---|
| A12 | Demo of DeCiFer basic command on prostate cancer patient A12 |
Recommendations and quality control
- We recommend to initially select a reasonably high maximum number of clusters with option
-K, e.g. a number that is ~2-3 times as large as(number of samples for the patient)+2. Further increase-Kif the selected best number of clusters is close to the maximum limit. - DeCiFer outputs the decreasing objective function which is used to select the number of clusters based on the Elbow criterion; if the function is still substantially decreasing near the selected maximum number of clusters please try to further increase this value.
- You can adapt the sensitivity of the Elbow criterion by adjusting the corresponding
--elbowparameter. In our experience, decreasing this value to ~0.002 gives better results for whole-genome sequences collected from ~5 samples from the same tumor; elbow values higher than this can result in overclustering with clusters containing visually distinct groups of DCF/CCF values. - Although DeCiFer performs model selection to select the best number of K clusters, given the specified range of clusters from
--minkto--maxkand the--elbowparameter, you can use the--printallkto print the output for all values of K specified. One output file is printed per value of K, each ending in_K#.tsvwhere#is the number of clusters. This can be useful to quickly explore how results may vary with different sensitivities, compared to re-running DeCiFer with different--elbowvalues. - If DeCiFer is taking more than a few days to run -- from having many SNVs, many samples, or both -- using fewer restarts (e.g.
--restarts 10) may be a sensible solution. We have observed very similar results for some datasets using a value of 100 and a lower value of 10. - Beta: use the
--conservativeCIsflag to have DeCiFer compute CIs that are wider and more conservative. This approach uses the DCF point values of the mutations assigned to a cluster to compute the cluster's CIs. Specifically, we compute the median of the distribution of DCF values (one for each mutation assigned to the cluster) and use bootstrap resampling to calculate CIs. To be conservative, we use the minimum and maximum observed median across 10,000 bootstrap replicates, instead of the medians that correspond to e.g. 0.025 and 0.975 quantiles.
Development
DeCiFer is in active development, please report any issue or question as this could help the development and improvement of DeCiFer. Known issues with current version are reported here below.
Contacts
DeCiFer has been developped and actively mantained by three previous Postdoctoral Research Associates at Princeton University in the research group of prof. Ben Raphael:
-
Simone Zaccaria (most of algorithmic core, likelihood computation, and Python implementation), group leader of the Computational Cancer Genomics research group at UCL Cancer Institute, London, UK
-
Gryte Satas (most of input/output and CF computations for single mutations), Postdoctoral Research Fellow at Memorial Sloan Kettering Cancer Center, NY, USA
-
Mohammed El-Kebir (generation of state trees), Assistant Professor in the Computer Science department at the University of Illinois at Urbana-Champaign (UIUC), IL, USA.
Additional active contributors to DeCiFer are:
- Brian Arnold, DataX Biomedical Data Scientist, Princeton University, USA.