Automation of the Kidney Function Prediction and Classification Through Ultrasound-based Kidney Imaging Using Deep Learning
This repository contains the code of the prediction model used in the paper Automation of the Kidney Function Prediction and Classification Through Ultrasound-based Kidney Imaging Using Deep Learning. The paper has been accepted by npj Digital Medicine. The propsed automated system ensembles 10 trained models to predict estimated glomerular filtration rate (eGFR), and 10 trained gradient-boosted tree models to classify CKD stage. Go to project blog post for more information. Open access of the paper on npj Digital Medicine
- 2019.03.19 Added GUI of the prediction module. See below for instructions.
This work uses Python 3.5.2. Before running the code, you have to install the following.
- torch==0.2.0_3
- torchvision==0.2.1
- pandas==0.20.3
- numpy==1.15.3
- xgboost==0.72
- opencv-python==3.4.0
- Pillow==5.1.0
The above dependencies can be installed using pip by running:
pip install -r requirement.txt
- Run get_models.sh to retrieve 10 trained CNN model weights and 10 trained XGBoost models. (Warning! The file size is about 1.7 GB).
bash get_models.sh
- Execute ensemble_predict.py
python3 ensemble_predict.py
- Input file path to a cropped kidney sonography. The images whould be 224 x 224 and cropped with tailor-cropped method mentioned in the paper.
use GPU: True
numbers of GPU: 1
Input image path:
- First time running will required loading the trained models, then the script shall return the predicted eGFR and the possibility of the subject's CKD stage being over stage III.
Use --help to see usage of ensemble_predict.py:
usage: ensemble_predict.py [-h] [-g]
optional arguments:
-h, --help show help message and exit
-g, --gpu_id assign GPU ID, default 0
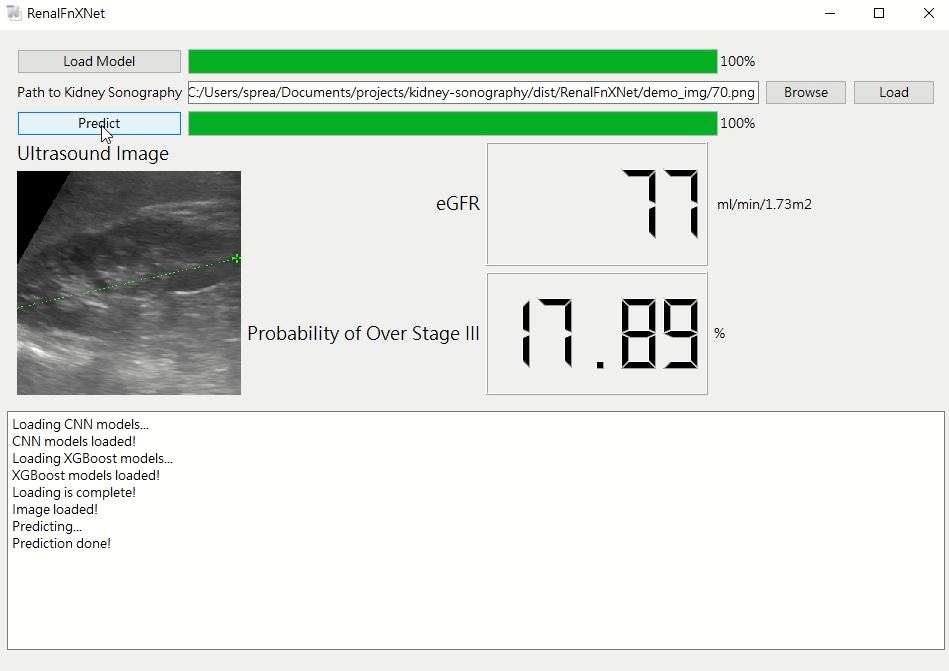
For ease of future clinical use, added a GUI for the predictiom module.
- Run get_models.sh to retrieve 10 trained CNN model weights and 10 trained XGBoost models. (Warning! The file size is about 1.7 GB).
bash get_models.sh
- Execute predict.py
python3 predict.py
Executable application for Windows environment. The full model version utilizes the 10-model-ensemble model used in our paper, it requires a GPU with 4GB VRAM. There is also a lite version with 5-model ensemble which requires 2GB VRAM, suitable for entry-level GPUs, yet at a cost of performance.
Dowload the file below and extract. Execute 'RenalFnXNet.exe'.
Full model version Requires GPU with 4GB of RAM
Lite version with 5-model ensembling Requires GPU with 2GB of RAM