A)
B)
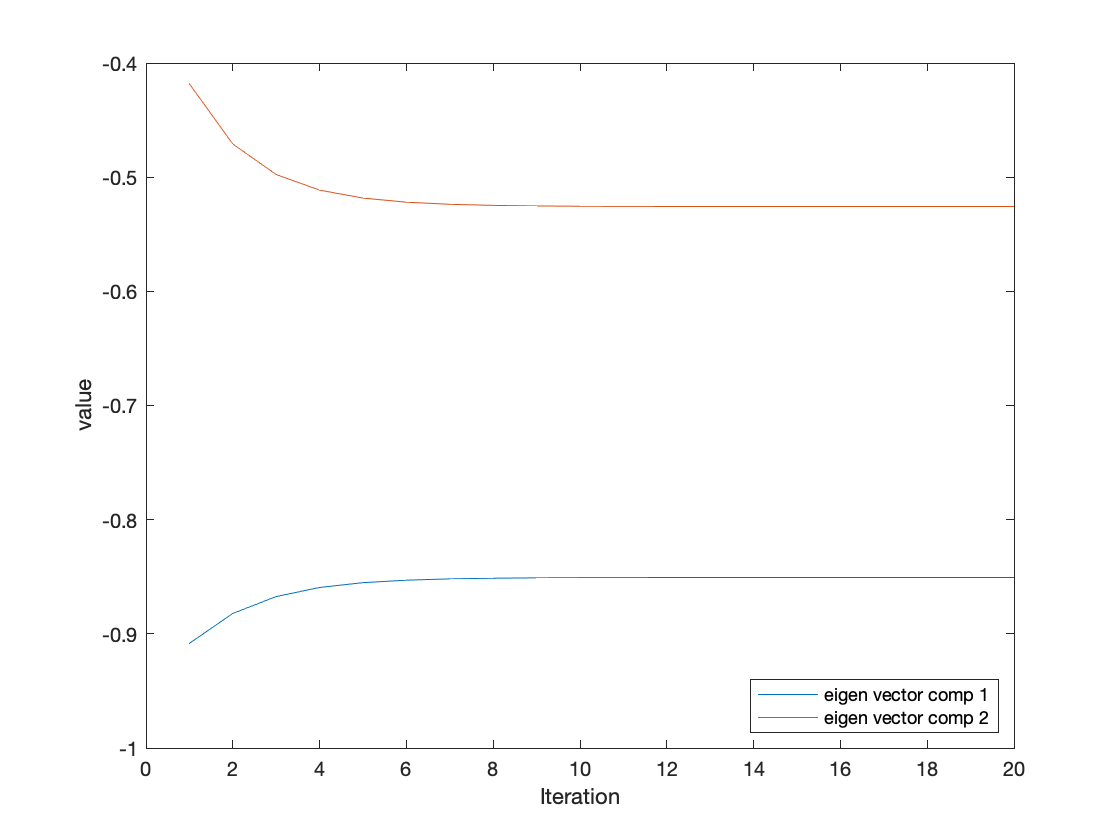
We obtain similar eigenvalues to the one obtained by matlab eig. For the eigenvector we had some initial diferences when it comes to the final values We were abled to obtain similar direction vectors but their individiual values are different. We ere able to rectify this by normalizing the vectors to unit norm. The script hw.m also includes the code for deriving the eigen vectors by "hand" using symbolic math solvers.
here we change the dataset to use the sample dataset two. Our primary change was to look into the 90th percentile in signal instead of the 99th percentile. It is clearly evident that in some channels a lot of the variability is no longer captured. Yet most of the trends are kept as one would expect from PCA.
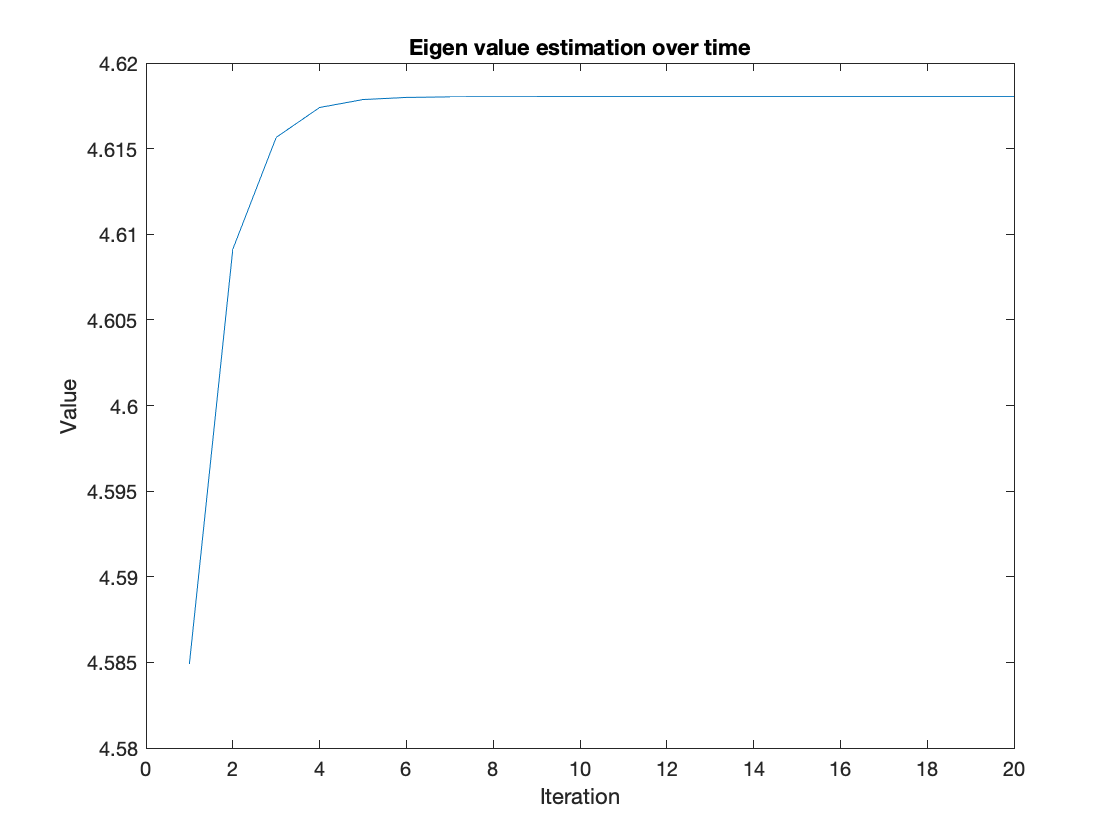
For thsi section we modified the iteration accunt to see how small the iteration count could get to reliably estimate the eigne value. Around the 20 iteration count we see stable levels of performance. This is a simpler signal and could perhaps not generalzie to larger matrices.
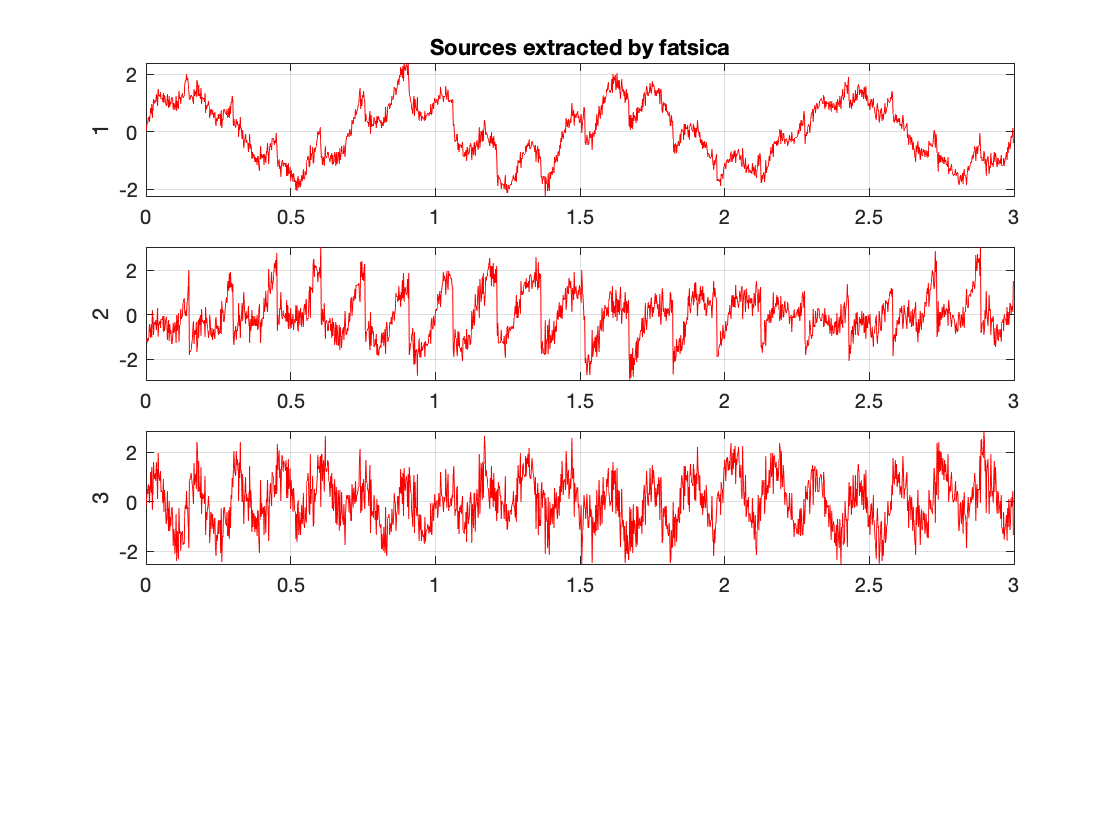
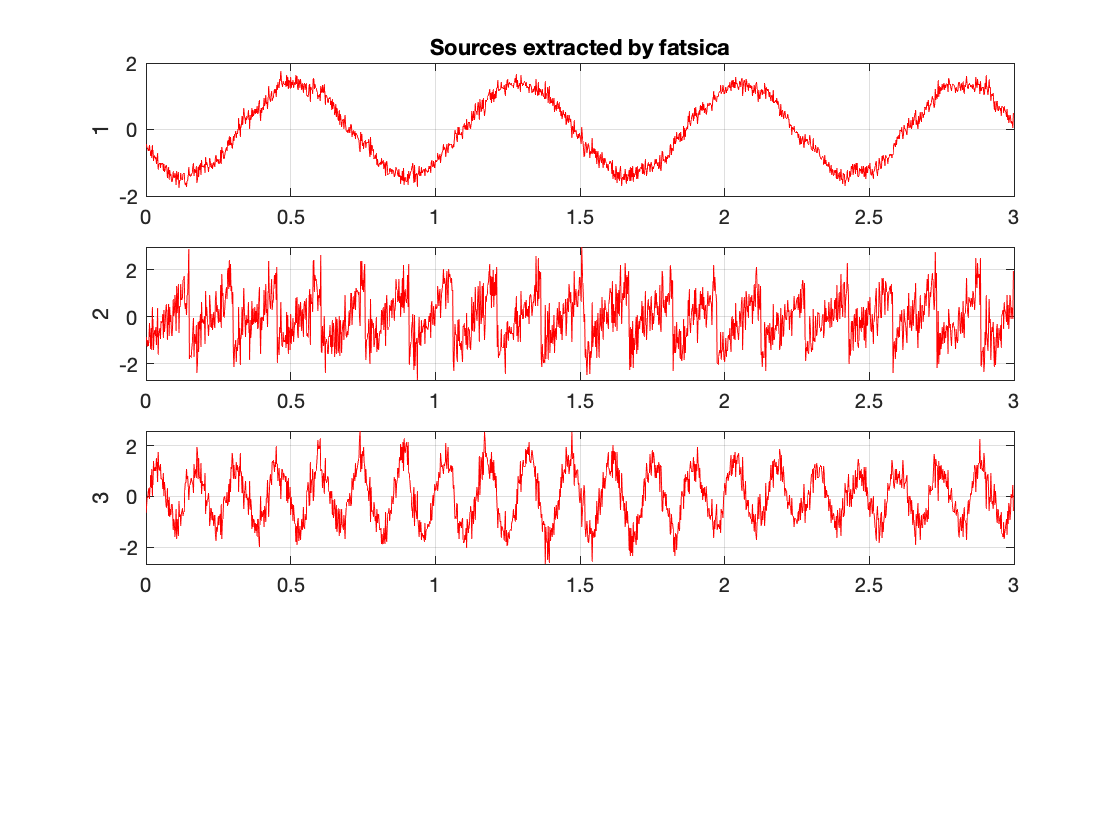
for this section we compared the changes caused by different appriximation approaches. We observed tanh,pow3, and skew to view a small subset of variability. We notice that tanh and po3 have the greatest agreeement. Meanwhile skew produces slightly different results. Bellow we have the skew and tanh example outputs of fastica.
noise removal utilizing NCSA seems to be more aggresive than using jade this is reflected in the summary statistics where the standard devieaiton of the signal is smallest with nsca. At a high level observation the signals appear to be rather similar. On closer inspection we can see differences in denoising ocmparing signals from channel 1 and channel 3 in the region with a rapid change seen in the 31 second mark we see that jade trails behind the NSCA method. Suggesting that jade is more sensitive to rapid changes in signals. Meanwhile for regions with a consistent signal( i.e flat) both methods are in agreement
this script appears to be using several model estimation techniques to fit a polynomial curve onto the the ECG signal. It allows for an interective interpretatatio nof the data which i find quite useful. Interestingly enough we can get an ok fit with just 2 parameters to be fit producing an error of 1.82% meanwhile for the
A comparison of PCA, KPCA, and ICA for dimensionality reduction in support vector machine
SVM was adopted "due to its remarkable characteristics such as good generalization performance, the absense of local minima and the sparse representation no solution" . SVMs are liked since they have the tendency to minimize the upper bound of the generalization error. Traditionally most applications would have used the raw feature space to do classification, but this is meant with a few limitations. Model performance is impacted by the correlated nature of the raw features. The scientific community therefore adopted several feature selection and/or extractions to minimize the number of unnecessary techniques. One series of feature extraction techniques that gained prominence was the dimensionality reduction techniques seen in PCA and KPCA. PCA using singular value decomposition produced a sieres of uncorrelated features. Meanwhile kernel PCA focuses on project the data onto a new space and applying linear PCA onto it. This allows KPCA to better generalize for non-linear features. ICA on the other hand focuses on identifying features that are statistically independent for each other. These different feature projection schemes result in varying feature sets, therefore, to validate their usefulness the researchers looked into 3 different datasets. They trained a SVM regression model of several openly available datasets. All 3 datasets had variables that provided a summary value of multiple events making their behavior nonlinear in most cases. It is for this reason that the best performing model was thus the KPCA followed by ICA. Although the data appeared to originate from different smaller sources which could have benefited from ICAs source separations abilities the top performer was still It is important to note that although KPCA was best performing it was also the slowest to run . PCA was the fastest model followed by ICA. Suggesting there are potential trade off considerations to be had when deciding which program to be used. ICA may not perform as well as KPCA but it’s quicker runtime would provide substantial benefit to biomedical applciaitons .