Compare similar sequences (*.fasta) to a reference (.fasta).
See the example report at: https://brwnj.github.io/idplot/
Nextflow is used to run the pipeline. Its installation instructions can be found at https://www.nextflow.io/ or installed via conda by way of bioconda. Bioconda includes a complete setup guide at https://bioconda.github.io/user/install.html.
Once your install completes and your channels are configured, run:
conda install nextflow
Executing the workflow using nextflow:
nextflow run brwnj/idplot -latest -with-docker \
--reference data/MN996532.fasta \
--fasta 'data/query_seqs/*.fasta'
This generally takes only a few minutes to complete which enables rapid screening for localized sequence similarity.
The reference sequence (--reference) should be a fasta with only one
sequence in it. Query sequences (--fasta) may either be single sequence
files or multi-sequence fasta files and you can specify more than one
using wildcards ('*').
Example sequences are found in data/query_sequences.
By default, output is written to ./results/idplot.html and can
be opened with an internet browser.
An example report is available at: https://brwnj.github.io/idplot/
In some cases it may be necessary to manually correct an alignment. In
this case, idplot can accept the alignment and skip its internal
alignment step. To do so, run:
nextflow run brwnj/idplot -latest -with-docker \
--alignment my_alignment_msa.fasta
The first sequence in the file will be used as the reference (root) sequence.
Options --reference and --fasta are both omitted in this case.
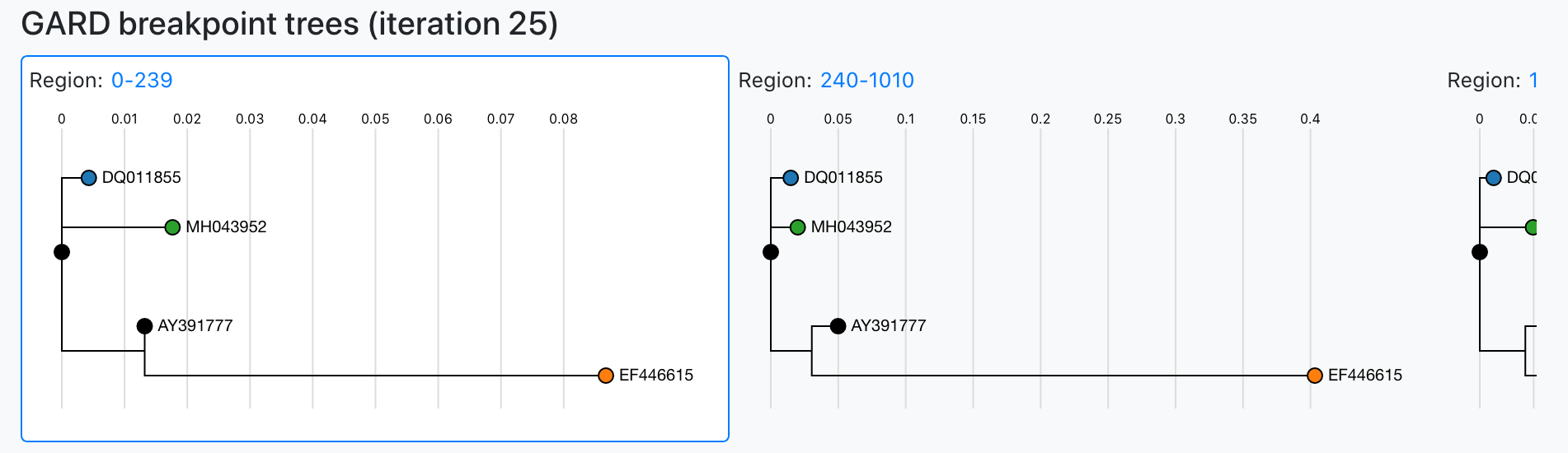
We have opted to employ GARD via HyPhy to identify breakpoints. For each GARD fit iteration, we pull the sequences for each breakpoint and build a tree using FastTree.
An example command enabling GARD with 12 MPI processes:
nextflow run brwnj/idplot -latest -with-docker \
--reference data/MN996532.fasta \
--fasta 'data/query_seqs/*.fasta' \
--gard --cpus 12
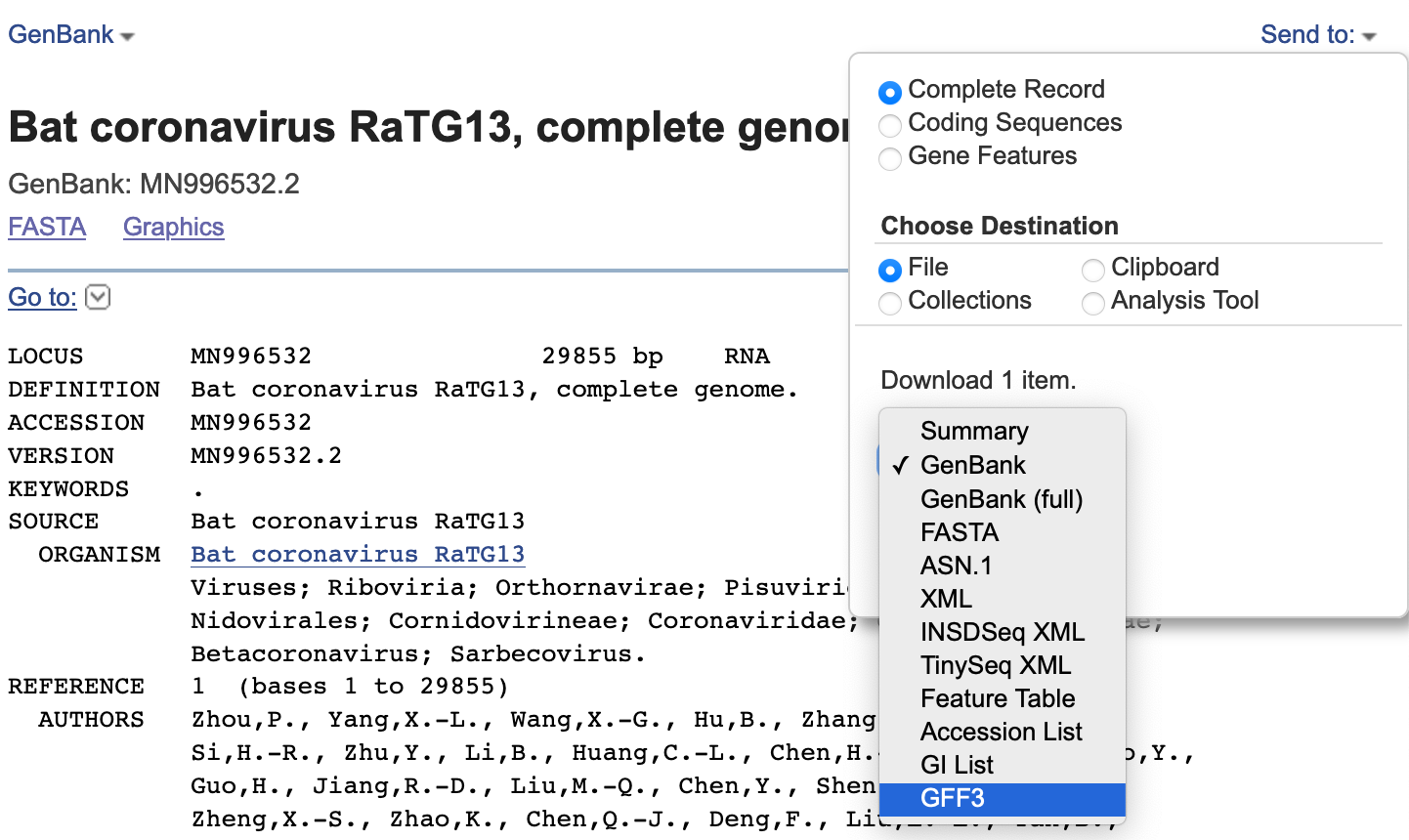
Publicly available reference sequences, like in NCBI, often have an accompanying annotation that can be included within the ANI plot. On NCBI, one can get a GFF from the reference page under Send to:
nextflow run brwnj/idplot -latest -with-docker \
--reference data/MN996532.fasta \
--fasta 'data/query_seqs/*.fasta' \
--gff MN996532.gff3
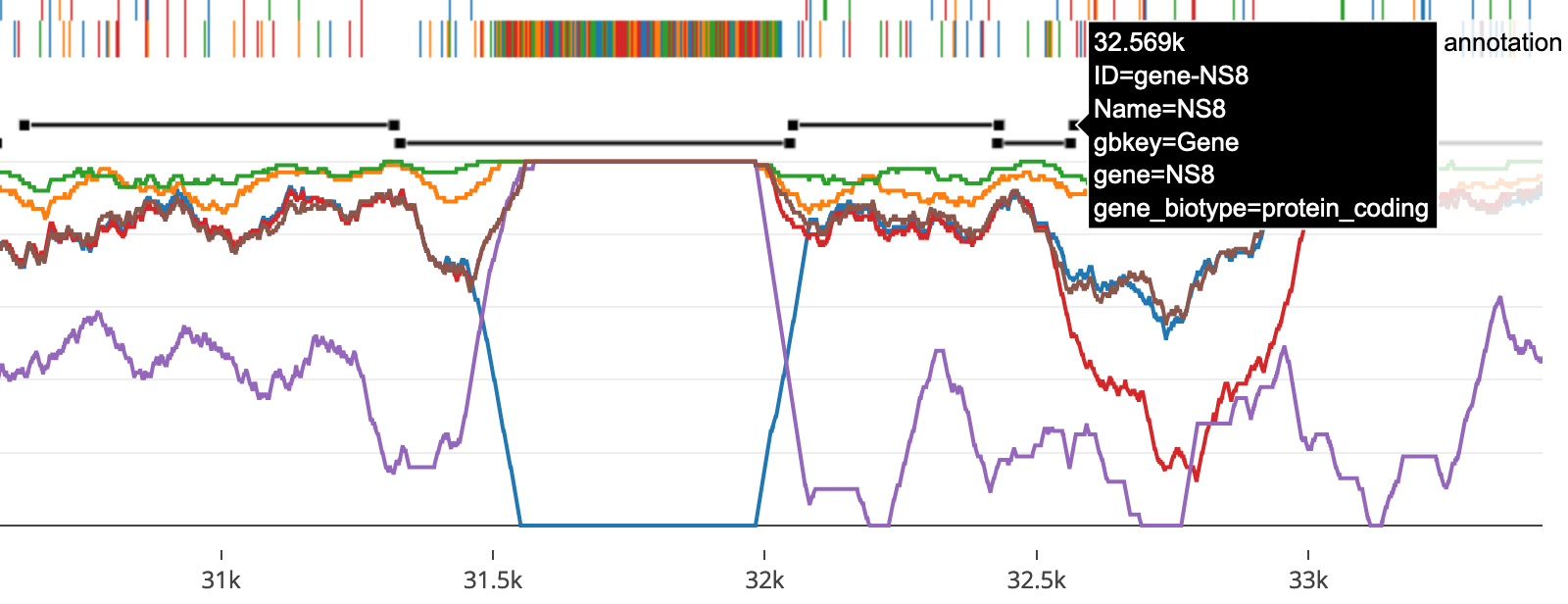
Within the report, this renders as:
As GFFs may have multiple feature types, we allow the reader to select their preferred feature type from the report header.
Coordinates in the original GFF will likely not match what is being displayed. Start and end coordinates are updated based on gaps introduced into the reference sequence during multiple sequence alignment.
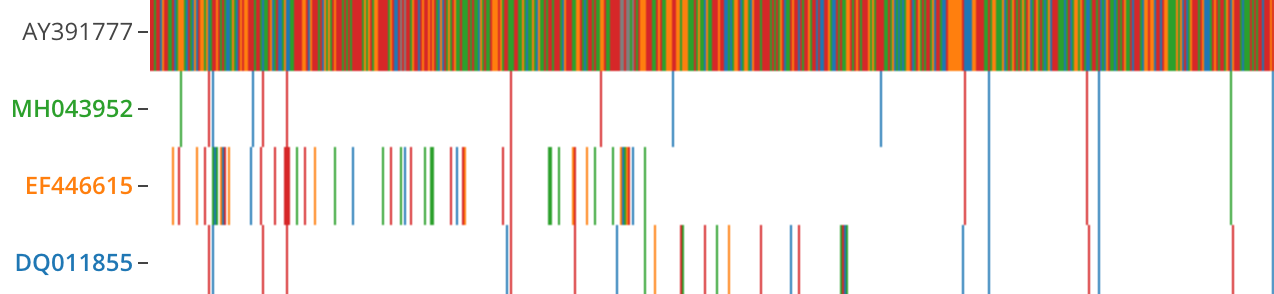
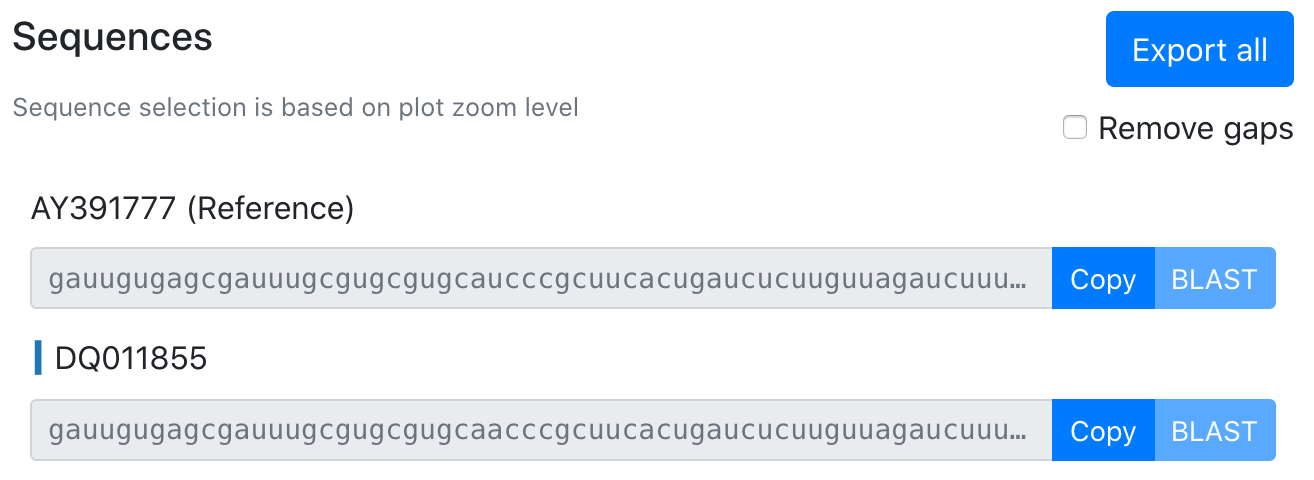
The reference sequence is fully colored in. Hovering along the reference shows the base for a given color.
Query sequences are colored at mismatches and gaps (gray).
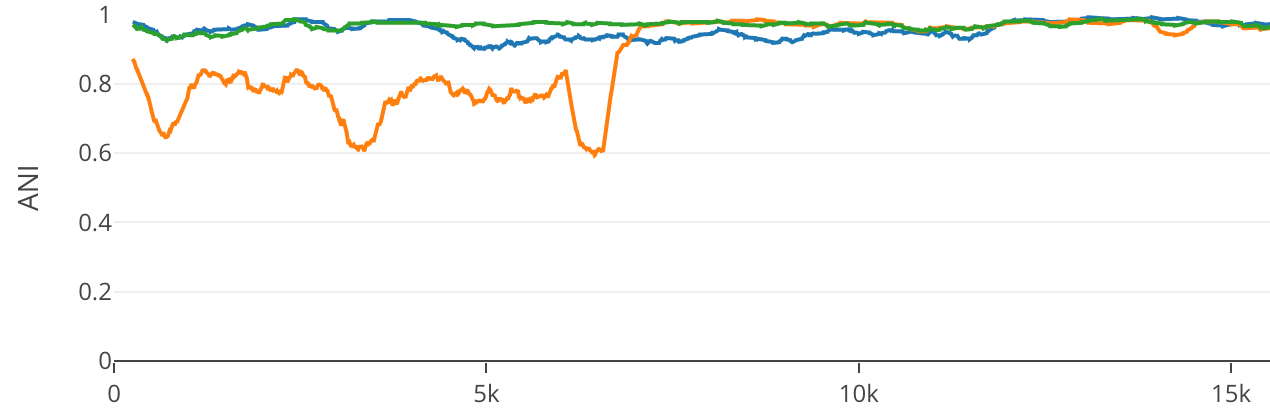
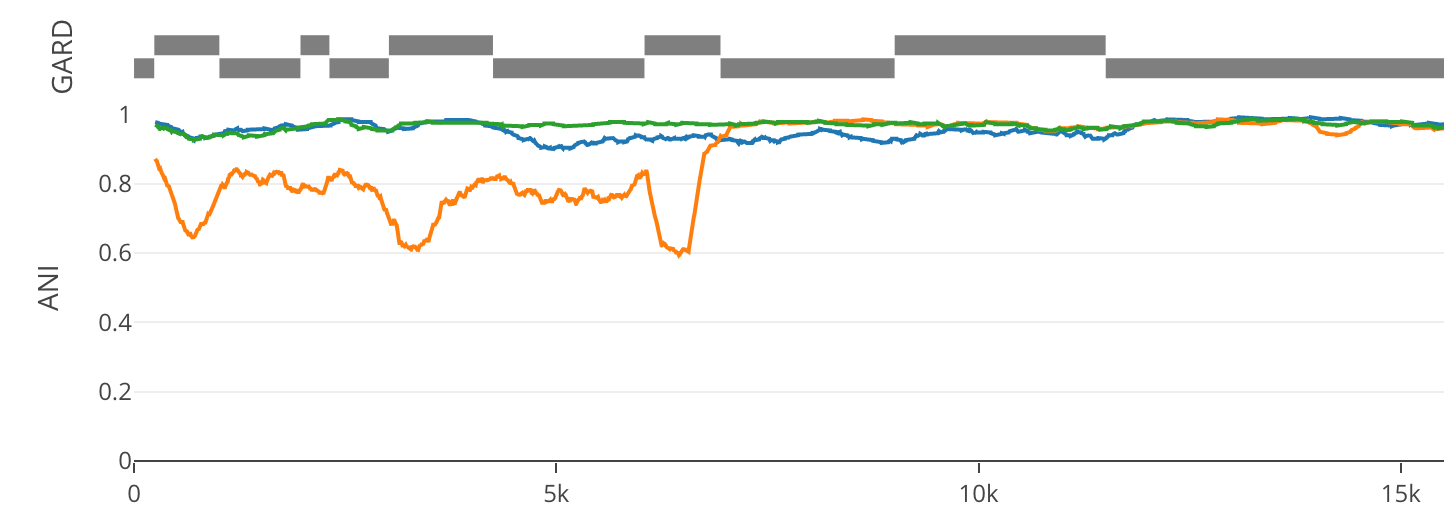
Percent ID is calculated across the window (default 500 bp) with the value being plotted at the center point. A 500 bp window will have 250 bp dead spots at the beginning and end of the reference length. No special treatment is given with respect to sequence content.
Sequence selection is based on the level of x-axis zoom of the plot. Sequence gaps can be removed using the toggle. The selected region can be copied to clipboard, sent directly to BLAST (when selection length is less than 8kb), or all sequences for a given region can be exported to FASTA.
Including --gard in your nextflow command adds breakpoint detection and updates available data and visualizations in the report.
Regions identified by GARD as breakpoints are highlighted between the MSA and ANI plots. Clicks on the regions will navigate to the respective dendrogram.
Per region dendrograms are generated using FastTree based off of regions identified by GARD.
Hovering over regions highlights the respective region in the GARD breakpoints track.
Clicking the region link will zoom the plot to facilitate downloading the sequence content for a given region.
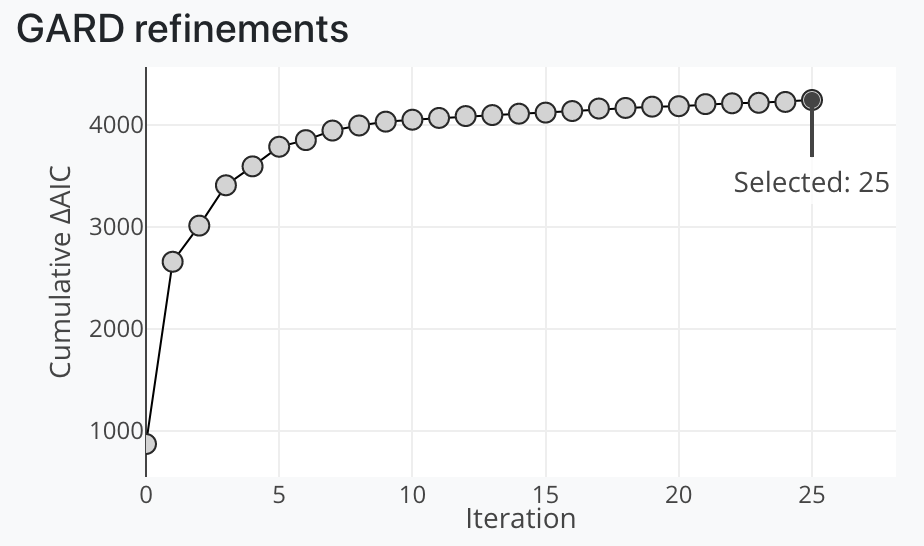
Breakpoints are identified over iterations by GARD, often to an unhelpful degree. This plot allows the user to explore breakpoints and trees across all GARD iterations. Selecting a new point will update the dendrograms and GARD breakpoints track.