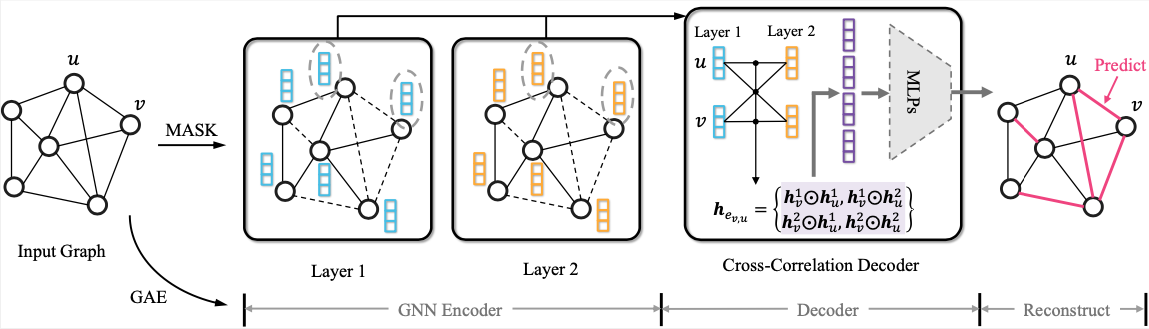
This is the PyG implementation for WSDM'23 paper: S2GAE: Self-Supervised Graph Autoencoders Are Generalizable Learners with Graph Masking
S2GAE is a generalized self-supervised graph representation learning method, which achieves competitive or better performance than existing state-of-the-art methods on different types of tasks including node classification, link prediction, graph classification, and molecular property prediction.
The required packages can be installed by running pip install -r requirements.txt.
Node classification (Cora, CiteSeer, and PubMed)
python s2gae_nc_acc.py --dataset `Cora`
Link prediction (ogbl-ddi, ogbl-collab, and ogbl-ppa)
python s2gae_large_lp.py --dataset "ogbl-ddi"
Graph classification
Node classification (Accuracy, %)
| Cora | CiteSeer | PubMed | A-Compute | A-Photo | Coauthor-CS | Coauthor-Physics | Ogbn-arxiv | Ogbn-proteins | |
|---|---|---|---|---|---|---|---|---|---|
| MVGRL | 85.86±0.15 | 73.18±0.22 | 84.86±0.31 | 88.70±0.24 | 92.15±0.20 | 92.87±0.13 | 95.35±0.08 | 68.33±0.31 | - |
| BGRL | 86.16±0.20 | 73.96±0.14 | 86.42±0.18 | 90.48±0.10 | 93.22±0.15 | 93.35±0.06 | 96.16±0.09 | 71.77±0.19 | _ |
| GraphMAE | 85.45±0.40 | 72.48±0.77 | 85.74±0.14 | 88.04±0.61 | 92.73±0.17 | 93.47±0.04 | 96.13±0.03 | 71.86±0.00 | 60.99±0.21 |
| MaskGAE | 87.31±0.05 | 75.20±0.07 | 86.56±0.26 | 90.52±0.04 | 93.33±0.14 | 92.31±0.05 | 95.79±0.02 | 70.99±0.12 | 61.23±0.19 |
| S2GAE(ours) | 86.15±0.25 | 74.60±0.06 | 86.91±0.28 | 90.94±0.08 | 93.61±0.10 | 91.70±0.08 | 95.82±0.03 | 72.02±0.05 | 63.33±0.12 |
Link prediction (AUC)
| Cora | CiteSeer | PubMed | Blogcatalog | Flickr | Ogbl-ddi | Ogbl-collab | Ogbl-ppa | |
|---|---|---|---|---|---|---|---|---|
| AUC | AUC | AUC | AUC | AUC | AUC | Hits@20 | Hits@50 | Hits@10 |
| GAE | 91.09±0.01 | 90.52±0.04 | 96.40±0.01 | 84.91±1.44 | 92.50±0.40 | 37.07±5.07 | 44.75±1.07 | 2.52±0.47 |
| GraphMAE | 89.19±0.00 | 91.20±0.11 | 93.72±0.00 | 76.60±1.32 | 88.69±0.04 | - | 22.79±1.62 | 0.18±0.28 |
| MaskGAE | 96.66±0.17 | 98.00±0.23 | 99.06±0.05 | 81.06±3.06 | 93.60±0.14 | 16.25±1.60 | 32.47±0.59 | 0.23±0.04 |
| S2GAE(ours) | 95.05±0.76 | 94.85±0.49 | 97.38±0.17 | 87.06±0.37 | 94.38±0.02 | 65.91±3.50 | 54.74±1.06 | 3.98±1.33 |
Graph classification (Accuracy, %)
| IMDB-B | IMDB-M | PROTEINS | COLLAB | MUTAG | REDDIT-B | NCI1 | |
|---|---|---|---|---|---|---|---|
| InfoGraph | 73.03±0.87 | 49.69±0.53 | 74.44±0.31 | 70.65±1.13 | 91.20±1.30 | - | 76.20±1.06 |
| GraphCL | 71.14±0.44 | 48.58±0.67 | 74.39±0.45 | 71.36±1.15 | 86.80±1.34 | 89.53±0.84 | 77.87±0.41 |
| MVGRL | 74.20±0.70 | 51.20±0.50 | - | - | 89.70±1.10 | 84.50±0.60 | - |
| GraphMAE | 75.52±0.66 | 51.63±0.52 | 75.30±0.39 | 80.32±0.46 | 88.19±1.26 | 88.01±0.19 | 80.40±0.30 |
| S2GAE(ours) | 75.76±0.62 | 51.79±0.36 | 76.37±0.43 | 81.02±0.53 | 88.26±0.76 | 87.83±0.27 | 80.80±0.24 |
If you find this work is helpful to your research, please consider citing our paper:
@inproceedings{tan2023s2gae,
title={S2GAE: Self-Supervised Graph Autoencoders Are Generalizable Learners with Graph Masking},
author={Tan, Qiaoyu and Liu, Ninghao and Huang, Xiao and Choi, Soo-Hyun and Li, Li and Chen, Rui and Hu, Xia},
booktitle={Proceedings of the 16th ACM International Conference on Web Search and Data Mining},
year={2023}
}
@article{tan2022mgae,
title={Mgae: Masked autoencoders for self-supervised learning on graphs},
author={Tan, Qiaoyu and Liu, Ninghao and Huang, Xiao and Chen, Rui and Choi, Soo-Hyun and Hu, Xia},
journal={arXiv preprint arXiv:2201.02534},
year={2022}
}