Domain adaptation based self-correction model for COVID-19 infection segmentation in CT images(https://doi.org/10.1016/j.eswa.2021.114848) accepted by Expert Systems with Applications
by Qiangguo Jin, Hui Cui, Changming Sun, Zhaopeng Meng, Leyi Wei, Ran Su.
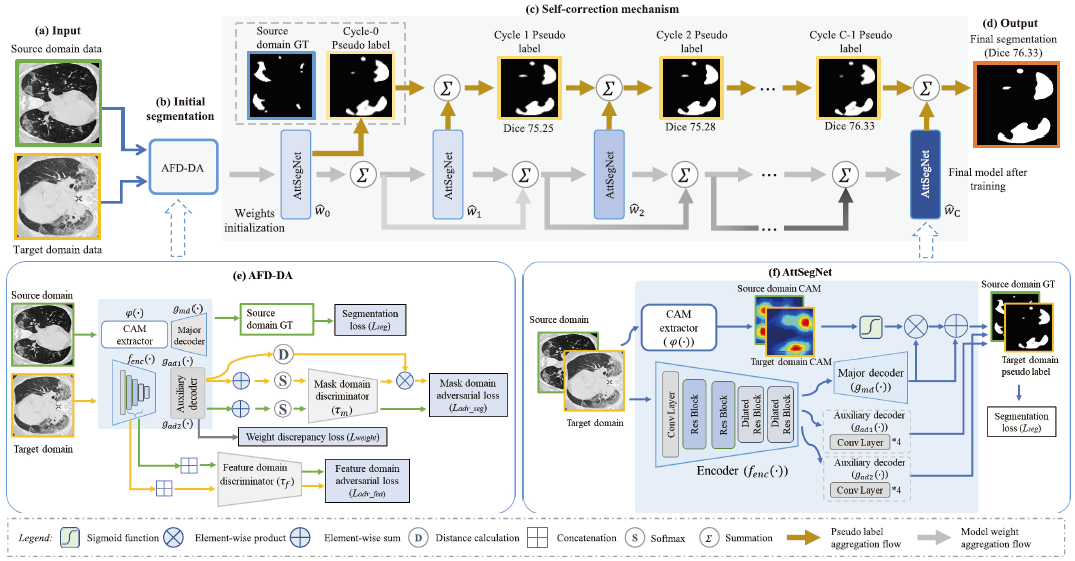
- Overview of the proposed DASC-Net for COVID-19 CT segmentation.
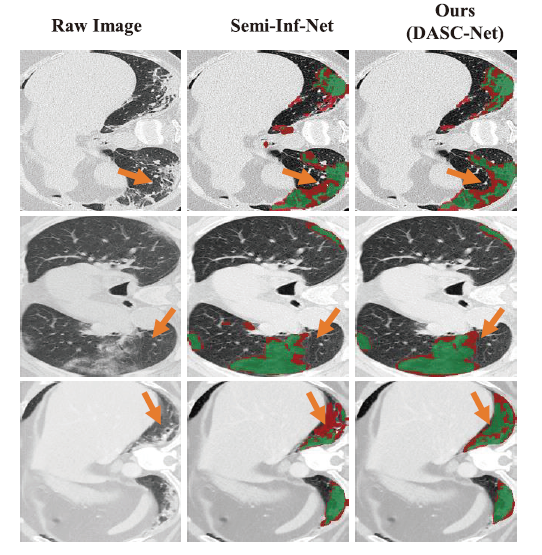
- Three examples of COVID-19 infection segmentation against Semi-Inf-Net. The segmentation results from Semi-Inf-Net were downloaded from the authors' GitHub repository(https://github.com/DengPingFan/Inf-Net) . The false predictions, i.e., false-positive and false-negative, are shown in red while the correct predictions are in green. The significant improvement by our model is marked with orange arrows.
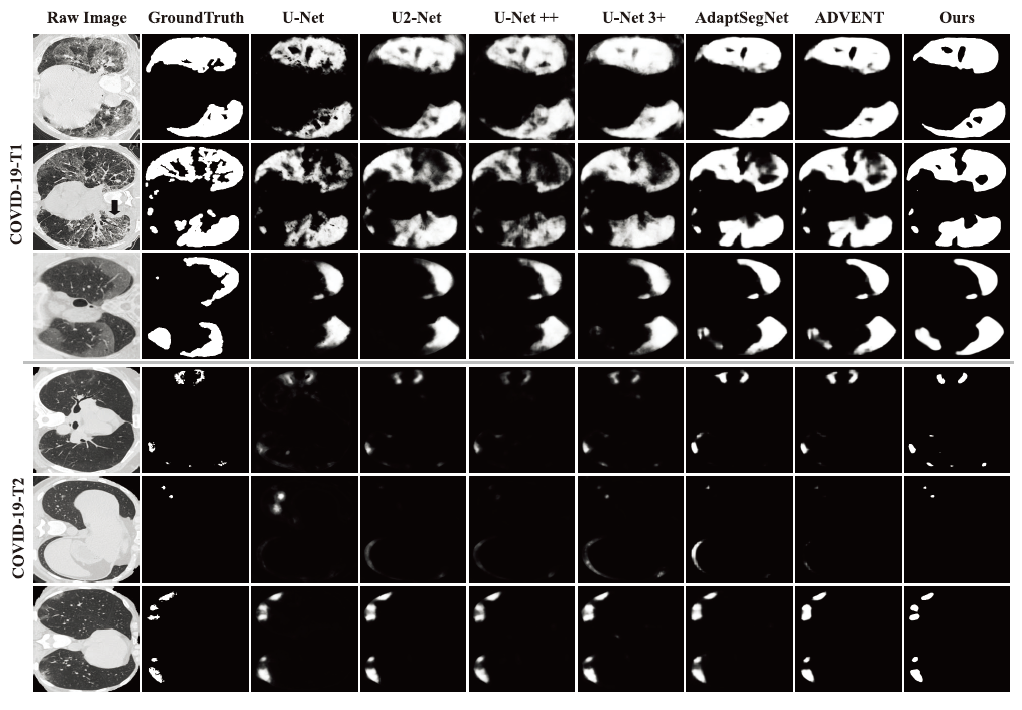
- Visual comparison of COVID-19 infection segmentation against other methods on two target datasets.
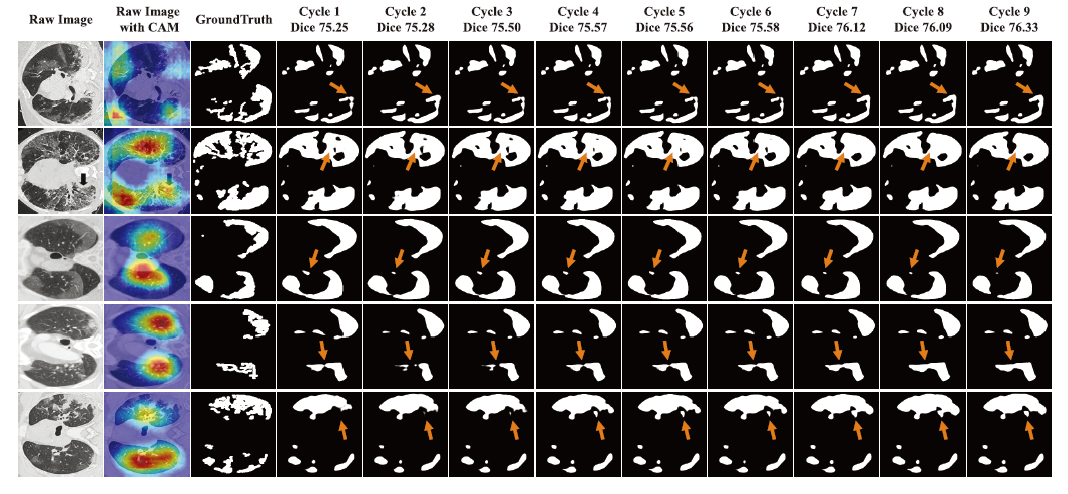
- Illustration of segmentation map in 9 cycles on COVID-19-T1. The refinement is marked with orange arrows.
- Releasing main code
- Detailed usage instruction
If the code is helpful for your research, please consider citing:
@article{JINDASC2021,
title = "Domain adaptation based self-correction model for COVID-19 infection segmentation in CT images",
author = "Qiangguo Jin, Hui Cui, Changming Sun, Zhaopeng Meng, Leyi Wei, and Ran Su",
journal = "Expert Systems with Applications",
year = "2021",
pages = "114848"
}
General questions, please contact 'qgking@tju.edu.cn'