Note
The newest code for Diffusion Curvature may be found here. This repository is not maintained, and is preserved only as a monument to the past. There have since been significant enhancements to the diffusion curvature algorithm.
Diffusion curvature is a pointwise extension of Ollivier-Ricci curvature, designed specifically for the often messy world of pointcloud data. Its advantages include:
- Unaffected by density fluctuations in data: it inherits the diffusion operator’s denoising properties.
- Fast, and scalable to millions of points: it depends only on matrix powering - no optimal transport required.
To install with pip (or better yet, poetry),
pip install diffusion-curvatureor
poetry add diffusion-curvatureConda releases are pending.
To compute diffusion curvature, first create a graphtools graph with
your data. Graphtools offers extensive support for different kernel
types (if creating from a pointcloud), and can also work with graphs in
the PyGSP format. We recommend using anistropy=1, and verifying that
the supplied knn value encompasses a reasonable portion of the graph.
from diffusion_curvature.datasets import torus
import graphtools
X_torus, torus_gaussian_curvature = torus(n=5000)
G_torus = graphtools.Graph(X_torus, anisotropy=1, knn=30)Graphtools offers many additional options. For large graphs, you can
speed up the powering of the diffusion matrix with landmarking: simply
pass n_landmarks=1000 (e.g) when creating the graphtools graph. If you
enable landmarking, diffusion-curvature will automatically use it.
Next, instantiate a
DiffusionCurvature
operator.
from diffusion_curvature.graphtools import DiffusionCurvature
DC = DiffusionCurvature(t=12)DiffusionCurvature (t:int, distance_type='PHATE', dimest=None, use_entropy:bool=False, **kwargs)
Initialize self. See help(type(self)) for accurate signature.
| Type | Default | Details | |
|---|---|---|---|
| t | int | Number of diffusion steps to use when measuring curvature. TODO: Heuristics | |
| distance_type | str | PHATE | |
| dimest | NoneType | None | Dimensionality estimator to use. If None, defaults to KNN with default params |
| use_entropy | bool | False | If true, uses KL Divergence instead of Wasserstein Distances. Faster, seems empirically as good, but less proven. |
| kwargs |
And, finally, pass your graph through it. The
DiffusionCurvature
operator will store everything it computes – the powered diffusion
matrix, the estimated manifold distances, and the curvatures – as
attributes of your graph. To get the curvatures, you can run G.ks.
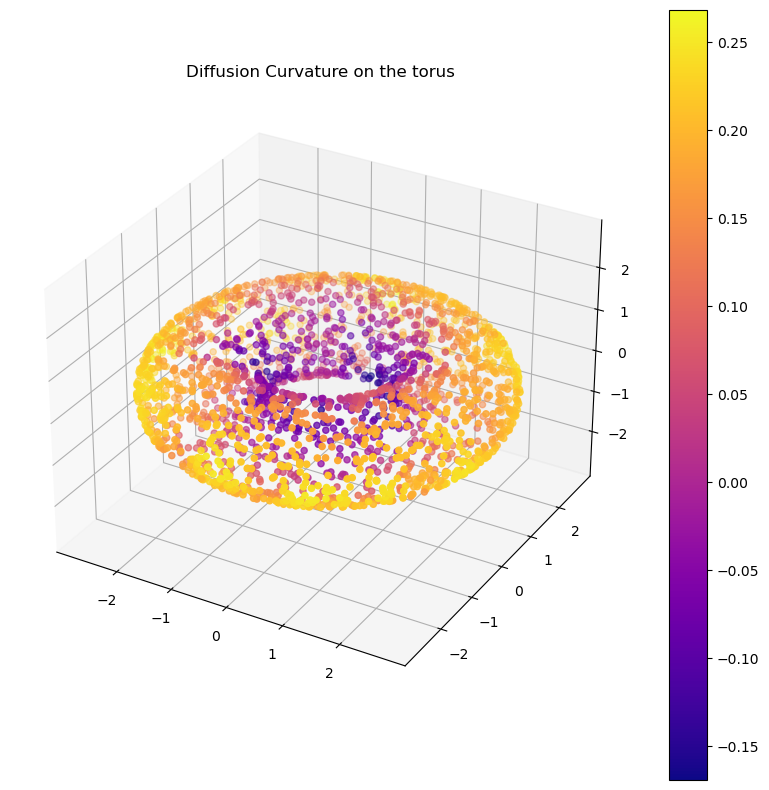
G_torus = DC.curvature(G_torus, dimension=2) # note: this is the intrinsic dimension of the dataplot_3d(X_torus, G_torus.ks, colorbar=True, title="Diffusion Curvature on the torus")If you have an adjacency matrix but no pointcloud, diffusion curvature may still be useful. The caveat, currently, is that our intrinsic dimension estimation doesn’t yet support graphs, so you’ll have to compute & provide the dimension yourself – if you want a signed curvature value.
If you’re only comparing relative magnitudes of curvature, you can skip this step.
For predefined graphs, we use our own
ManifoldGraph
class. You can create one straight from an adjacency matrix:
from diffusion_curvature.manifold_graph import ManifoldGraph, diffusion_curvature, diffusion_entropy_curvature, entropy_of_diffusion, wasserstein_spread_of_diffusion, power_diffusion_matrix, phate_distances
from diffusion_curvature.kernels import gaussian_kernel
import numpy as np# if you want (or have) to compute your own A
A = gaussian_kernel(X_torus, kernel_type="adaptive", k = 20, anisotropic_density_normalization=1)
np.fill_diagonal(A,0)
# initialize the manifold graph; input your computed dimension along with the adjacency matrix
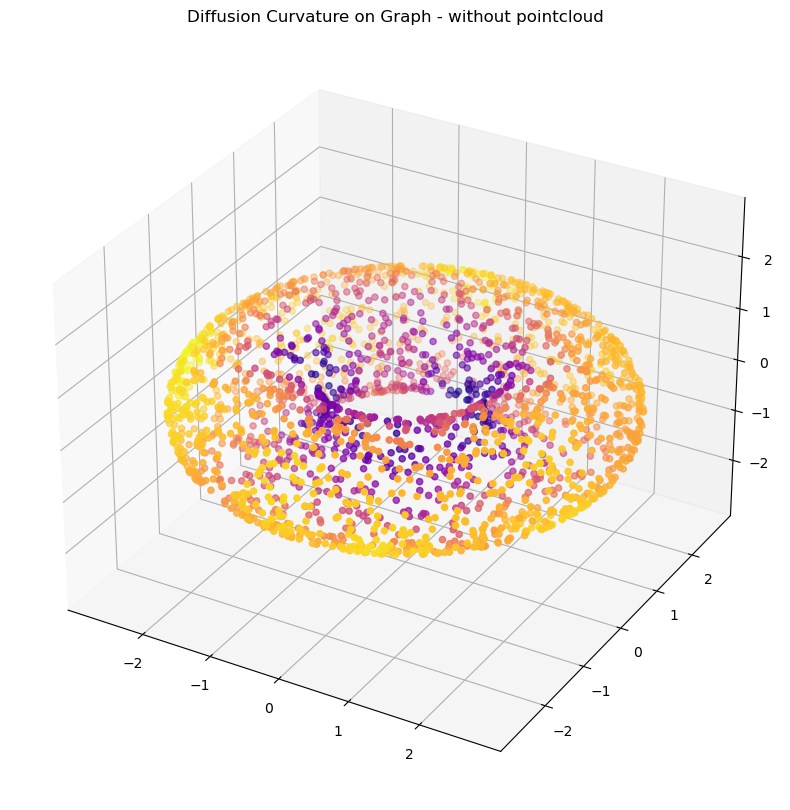
G_pure = ManifoldGraph(A = A, dimension=2, anisotropic_density_normalization=1)G_pure = diffusion_curvature(G_pure, t=8)
plot_3d(X_torus, G_pure.ks, title = "Diffusion Curvature on Graph - without pointcloud")Alternately, to compute just the relative magnitudes of the pointwise
curvatures (without signs), we can directly use either the
wasserstein_spread_of_diffusion
(which computes the entropy_of_diffusion
function (which computes the entropy of each t-step diffusion). The
latter is nice when the manifold’s geodesic distances are hard to
estimate – it corresponds to replacing the wasserstein distance with the
KL divergence.
Both of these estimate an “inverse laziness” value that is inversely proportional to curvature. To use magnitude estimations in which the higher the curvature, the higher the value, we can simply take the reciprocal of the output.
# for the wasserstein version, we need manifold distances
G_pure = power_diffusion_matrix(G_pure,t=8)
G_pure = phate_distances(G_pure)
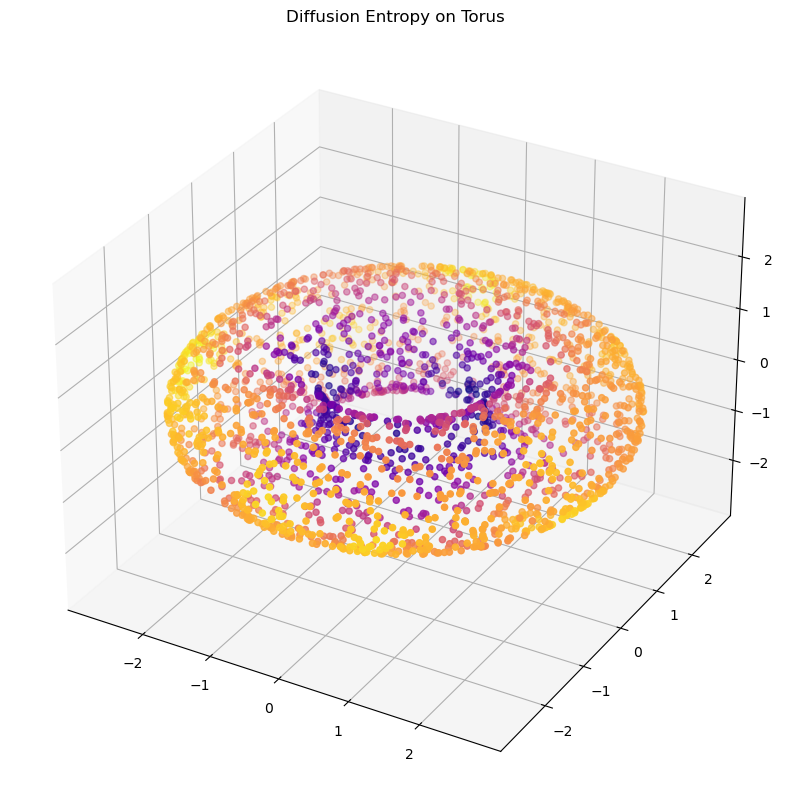
ks_wasserstein = wasserstein_spread_of_diffusion(G_pure)# for the entropic version, we need only power the diffusion operator
G_pure = power_diffusion_matrix(G_pure, t=8)
ks_entropy = entropy_of_diffusion(G_pure)plot_3d(X_torus, 1/ks_entropy, title="Diffusion Entropy on Torus")