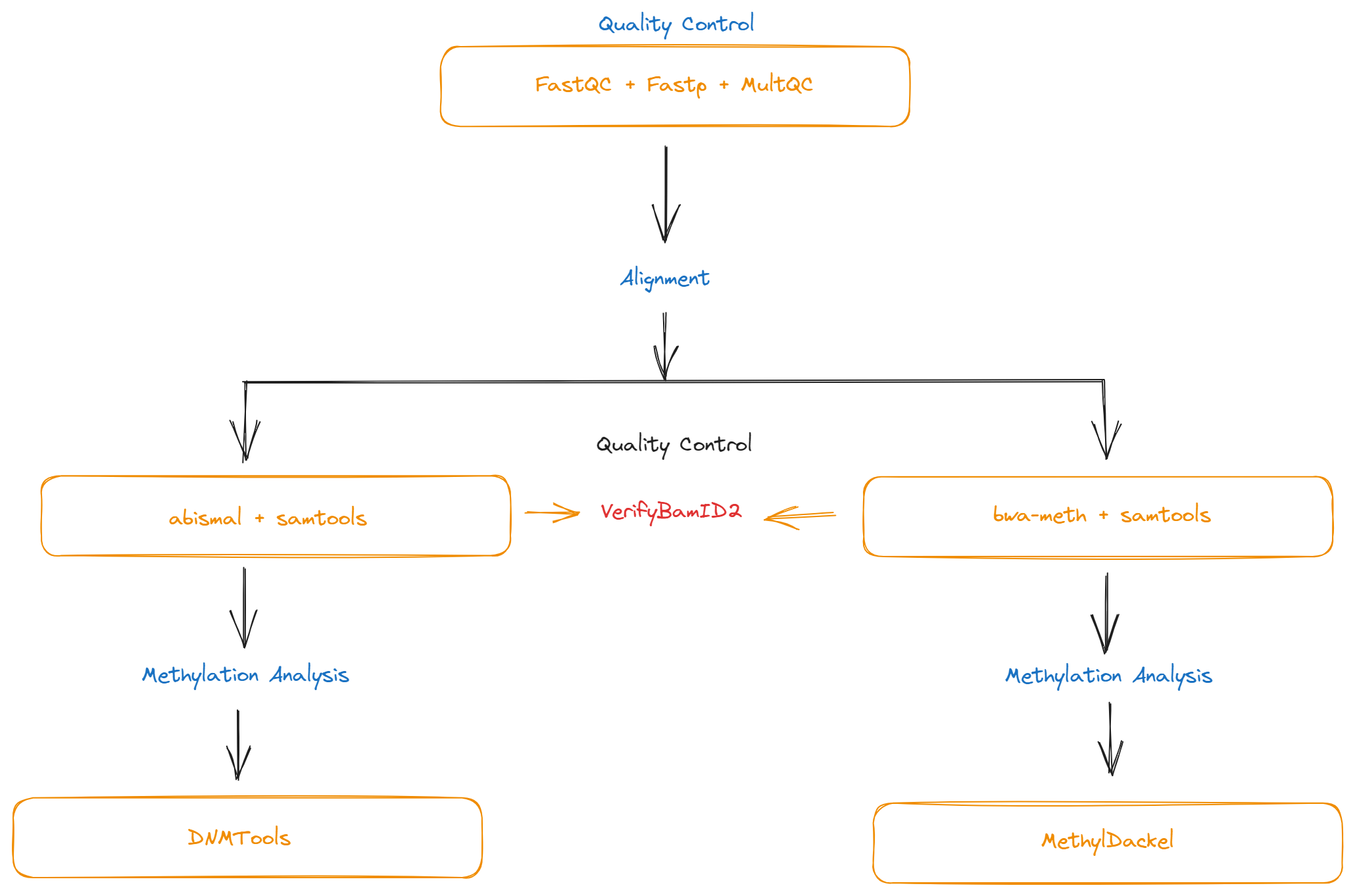
A Snakemake workflow that enables data processing of methylation sequencing platforms e.g bisulfite sequencing and enzymatic-methyl sequencing.
- FastQC
- Fastp
- MultiQC
- Two aligners are used here;
- VerifyBamID2
Two methods are employed;
- MethylDackel
- Integrates well with bwa-meth
- DMNTools
- Integrates well with abismal
<path-to-output-folder>\
| <project-name>\qc\
| fastqc\
| <sample>_<read>_fastqc.html
| <sample>_<read>_fastqc.zip
| fastp\
| <sample>_<read>.trimmed_fastqc.html
| <sample>_<read>.trimmed_fastqc.html
| <sample>_1.trimmed.fastq
| <sample>_2.trimmed.fastq
| <sample>_u1.fastq
| <sample>_u2.fastq
| <sample>.merged.fastq
| <sample>.failed.fastq
| <sample>.html
| <sample>.json
| multiqc\
| trimmed\
| multiqc_data\
| multiqc_report.html
| untrimmed\
| multiqc_data\
| multiqc_report.html
1. bwa-meth + samtools + VerifyBamID2
<path-to-output-folder>\
| <project-name>\alignment\bwa\
| unsorted/<sample>.sam
| sorted/
| <sample>.bam
| <sample>.bai
| <sample>.ancestry
| <sample>.selfsm
| /picard
| <sample>.bam
| <sample>.bam.bai
| <sample>.metrics.txt
2. abismal + samtools + VerifyBamID2
<path-to-output-folder>\
| <project-name>\alignment\abismal
| <sample>.bam
| <sample>.sorted.bam
| <sample>.sorted.bai
| <sample>.filtered.bam
| stats\
| <sample>.metrics.yaml
| <sample>.filtered.metrics.yaml
| verify_bam_id\
| <sample>.ancestry
| <sample>.selfsm
1. DNMTools
<path-to-output-folder>\
| <project-name>\methyl\dnmtools\
| <sample>.bsrate
| <sample>_single_base.meth
| <sample>_symmetric.meth
| <sample>_global.meth
| <sample>.hmr
| <sample>.hypermr
| <sample>.epiread
| <sample>.entropy.meth
| <sample>.avg.meth
2. MethylDackel - To be completed
- Genome (hg38)
- Canonical cis-Regulatory elements
Set-up working conda environment conda create --name <envname> --file requirements.txt
NB:
- The above step asumes you have conda already installed.
- This workflow was built on Ubuntu20.04. Other platforms were not tested.
launch the pipeline form within the <snakemake-methylseq>\<workflow> directory
All Rules: bash run_test.sh all
-
Quality Control
bash run.sh qc -
Alignment
- bwa-meth:
bash run.sh alignment bwa - abismal:
bash run.sh alignment abismal
- bwa-meth:
-
Methylation Analysis
- DNMTools:
bash run.sh methyl dnmtools - MethylDackel:
bash run.sh methyl methyldackel
- DNMTools:
-
Quality Control
bash run.sh qc - <num_cores> -
Alignment
- bwa-meth:
bash run.sh alignment bwa - abismal:
bash run.sh alignment abismal
- bwa-meth:
-
Methylation Analysis
- DNMTools:
bash run.sh methyl dnmtools - MethylDackel:
bash run.sh methyl methyldackel
- DNMTools:
- Add script to download and index reference files
- Add script to download
svd_mufor VerifyBamID2 - Fix MethylDackel environment for to complete analysis
- Work on the format conversion between abismal and bwa-meth
- Fix errors from rules
entropyandavg_meth_level_region - Provide PC plots for contamination of samples
- Add notes on updating the config file
- Add documentation on rules and their purpose