Several MIT courses involving numerical computation, including 18.06, 18.303, 18.330, 18.335/6.337, and 18.337/6.338, are beginning to use Julia, a fairly new language for technical computing. This page is intended to supplement the Julia documentation with some simple tutorials on installing and using Julia targeted at MIT students. See also our Julia cheatsheet listing a few basic commands, as well as the Learn Julia in Y minutes tutorial page.
In particular, we will be using Julia in the IJulia browser-based enviroment, which leverages your web browser and IPython to provide a rich environment combining code, graphics, formatted text, and even equations, with sophisticated plots via Matplotlib.
See also Viral Shah's Julia slides and the January 2013 Julia Tutorial videos.
Traditionally, these sorts of courses at MIT have used Matlab, a high-level environment for numerical computation. Other possibilities might be Scientific Python (the Python language plus numerical libraries), GNU R, and many others. Julia is another high-level free/open-source language for numerical computing in the same spirit, with a rich set of built-in types and libraries for working with linear algebra and other types of computations, with a syntax that is superficially reminiscent of Matlab's.
High-level dynamic programming languages (as opposed to low-level languages like C or static languages like Java) are essential for interactive exploration of computational science. They allow you to play with matrices, computations on large datasets, plots, and so on without having to worry about managing memory, declaring types, or other minutiae—you can open up a window and start typing commands to immediately get results.
The traditional problem with high-level dynamic languages, however, is that they are slow: as soon as you run into a problem that cannot easily be expressed in terms of built-in library functions operating on large blocks of data ("vectorized" code), you find that your code is suddenly orders of magnitude slower that equivalent code in a low-level language. The typical solution has been to switch to another language (e.g. C or Fortran) to write key computational kernels, calling these from the high-level language (e.g. Matlab or Python) as needed, but this is vastly more difficult than writing code purely in a high-level language and imposes a steep barrier on anyone hoping to transition from casual experimentation to "serious" numerical computation. Julia mostly eliminates this issue, because it is carefully designed to exploit a "just-in-time compiler" called LLVM, making it possible to write high-level code in Julia that achieves near-C speed. (It also means that we can perform meaningful performance experiments easily in courses where this matters, e.g. in 18.335.)
Speed, while avoiding the "two-language" problem of requiring C or Fortran for critical code, is the initial draw of Julia, but there are a few other nice points. Unlike Matlab, it is free/open-source software, which eliminates licensing headaches and allows you to look inside the Julia implementation to see how it works (since Julia is mostly written in Julia, its code is much more readable than a language like Python that is largely implemented in low-level C). For calling existing code, it has easy facilities to call external C or Fortran libraries or to call Python libraries. Multiple dispatch makes it especially easy to overload operations and functions for new types (e.g. to add new vector or numeric types). Julia's built-in metaprogramming facilities make it easy to write code that generates other code, essentially allowing you to extend the language as needed. And so on...
Note: If you have MIT web certificates, you may be able to run IJulia remotely on our cluster, without installing anything, by going to https://ijulia.csail.mit.edu:8000. We are still setting up this server and availability may currently be intermittent, however.
First, install IPython and related scientific-Python packages (SciPy and Matplotlib). The simplest way to do this on Mac and Windows is by downloading the Anaconda package and running its installer.
- Important: on Windows, the Anaconda installer window gives options Add Anaconda to the System Path and also Register Anaconda as default Python version of the system. Be sure to check these boxes.
Second, download Julia version
0.2 and run the installer. Do not
download version 0.1. Then run the Julia application (double-click on
it); a window with a julia> prompt will appear. At the prompt,
type Pkg.add("Homebrew") on MacOS or Pkg.add("RPMmd") on Windows (or nothing on Linux) and then type:
Pkg.add("IJulia")
Pkg.add("PyPlot")
- If you ran into a problem with the above steps, after fixing the
problem you can type
Pkg.build()to try to rerun the install scripts. - If you tried it a while ago, try running
Pkg.update()and try again: this will fetch the latest versions of the Julia packages in case the problem you saw was fixed. RunPkg.build("IJulia")if your Julia version may have changed. If this doesn't work, try just deleting the whole.juliadirectory in your home directory (on Windows, it is calledAppData\Roaming\julia\packagesin your home directory) and re-adding the packages. - On MacOS, you currently need MacOS 10.7 or later; MacOS 10.6 doesn't work (unless you compile Julia yourself, from source code).
- If
Pkg.add("IJulia")says that the IJulia package doesn't exist, then you probably downloaded Julia 0.1 by mistake. Download Julia 0.2, runPkg.update()in Julia, and try again. - On Windows, if you get an error
no module named sitewhenusing PyPlot, probably you forgot to check the boxes in the Anaconda installer (above) to register Anaconda as the default Python version. Either re-install Anaconda or set the environment variablesPYTHONHOME=C:\AnacondaandPYTHONPATH=C:\Anaconda\Lib. - If the browser opens the notebook and
1+1works but basic functions likesin(3)don't work, then probably you are running Python and not Julia. Look in the upper-left corner of the notebook window: if it says IP[y]: Notebook then you are running Python. Probably this was because yourPkg.add("IJulia")failed and you ignored the error. - Internet Explorer 8 (the default in Windows 7) or 9 don't work with the notebook; use Firefox (6 or later) or Chrome (13 or later). Internet Explorer 10 in Windows 8 works (albeit with a few rendering glitches), but Chrome or Firefox is better.
- If the notebook opens up, but doesn't respond (the input label is
In[*]indefinitely), try runningipython notebook(without Julia) to see if1+1works in Python. If it is the same problem, then probably you have a firewall running on your machine (this is common on Windows) and you need to disable the firewall or at least to allow the IP address 127.0.0.1. (For the Sophos endpoint security software, go to "Configure Anti-Virus and HIPS", select "Authorization" and then "Websites", and add 127.0.0.1 to "Authorized websites"; finally, restart your computer.) - On Windows, if the notebook says the kernel crashed (repeatedly) and there is an error message
"julia-readline.exe" is not recognizedin the command-line window, the problem is that you aren't running theipythoncommand from within the Juliabindirectory; see the "Important" note below.
Julia is also installed on MIT's Athena Computing
Environment. Any MIT student can use the
computers in the Athena Clusters
on campus, and you can also log in remotely to athena.dialup.mit.edu
via ssh. (Note: the
linux.mit.edu dialup does not work with Julia.)
In the terminal of an Athena machine, type:
add -f julia_v0.2
export PYTHONPATH=/mit/julia_v0.2/lib/python2.7/site-packages
to load the Julia and IPython software locker.
The first time you use Julia on Athena, you will need to set up IJulia: run julia, and at the julia> prompt, type
Pkg.update()
Pkg.add("PyPlot")
Pkg.add("IJulia")
Thereafter, you can open the IJulia notebook in the web browser by
running ipython notebook --profile julia as described below.
Note: You can only run the IJulia notebook if you physically go to an
Athena cluster. If you are logging in remotely to
athena.dialup.mit.edu, you can type julia to use the text terminal. You can still use PyPlot graphics via dialup, but in order to do so you will need to set up X Windows on your computer.
Julia is improving rapidly, so it won't be long before you want to update to a more recent version. The same is true of Julia add-on packages like PyPlot. To update the packages only, keeping Julia itself the same, just run:
Pkg.update()
at the Julia prompt (or in IJulia).
If you download and install a new version of Julia from the Julia web
site, you will also probably want to update the packages with
Pkg.update() (in case newer versions of the packages are required
for the most recent Julia). In any case, if you install a new Julia
binary (or do anything that changes the location of Julia on your
computer), you must update the IJulia installation (to tell IPython
where to find the new Julia) by running Pkg.build("IJulia") at the
Julia command line (not in IJulia).
Once you have followed the installation steps above, open up the command line (the Terminal program in MacOS or the Command Prompt on Windows) and type:
ipython notebook --profile julia
- Important: On Windows, you must run the above command from the
binsubdirectory of the Julia directory that you downloaded/unpacked. e.g., open up the window for thebinsubdirectory and then choose Open Command Prompt from the File menu (in Windows 8) in order to type the above command.
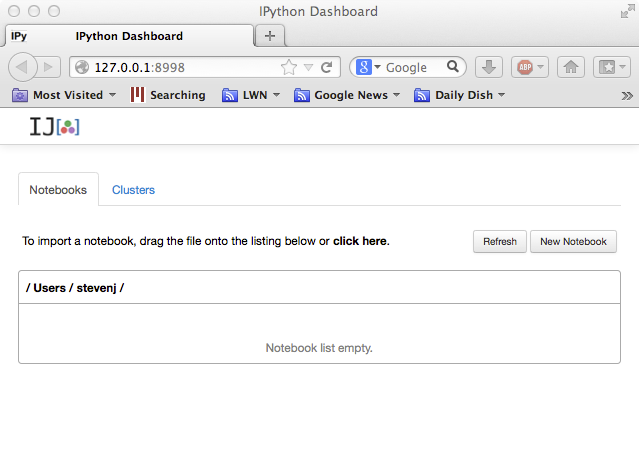
A "dashboard" window like this should open in your web browser:
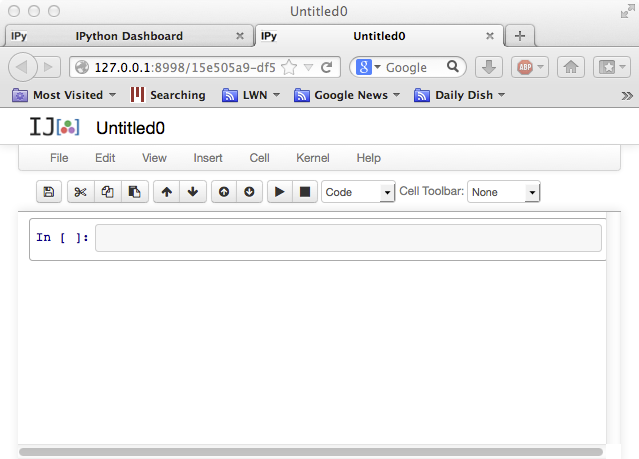
Now, click on the New Notebook button to start a new "notebook". A notebook will combine code, computed results, formatted text, and images; for example, you might use one notebook for each problem set. The notebook window that opens will look something like:
You can click the "Untitled0" at the top to change the name, e.g. to
"My first Julia notebook". You can enter Julia code at the In[ ]
prompt, and hit shift-return to execute it and see the results.
If you hit return without the shift key, it will add additional
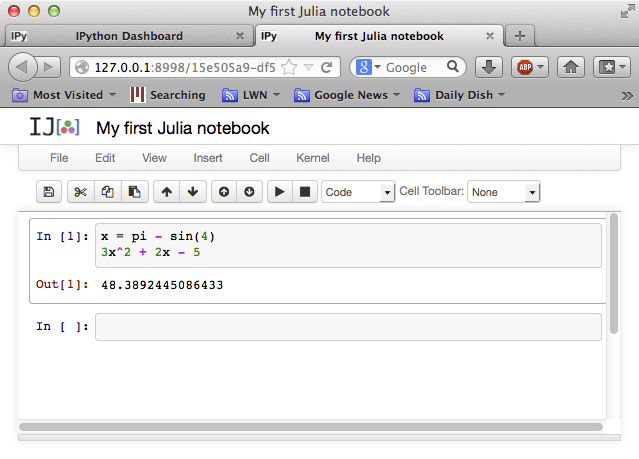
lines to a single input cell. For example, we can define a variable
x (using the built-in constant pi and the built-in function
sin), and then evaluate a polynomial 3x^2 + 2x - 5 in terms of x
(note that, unlike Matlab or Python, we don't have to type 3*x^2 if
we don't want to: a number followed by a variable is automatically
interpreted as multiplication without having to type *):
The result that is printed (in Out[1]) is the last expression from
the input cell, i.e. the polynomial. If you want to see the value of
x, for example, you could simply type x at the second In[ ] prompt
and hit shift-return.
See, for example, the mathematical operations in the Julia manual for many more basic math functions.
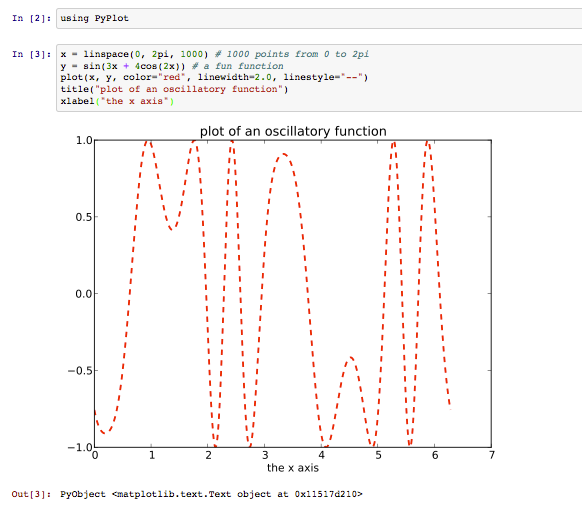
There are several plotting packages available for Julia. If you followed the installation instructions, above, you already have one full-featured Matlab-like plotting package installed: PyPlot, which is simply a wrapper around Python's amazing Matplotlib library.
To start using PyPlot to make plots in Julia, first type using PyPlot at an input prompt and hit shift-enter. using is the Julia
command to load an external
module (which
must usually be
installed
first, e.g. by the Pkg.add("PyPlot") command from the installation
instructions above).
Then, you can type any of the commands from Matplotlib, which includes equivalents for most of the Matlab plotting functions. For example:
Currently, printing a notebook from the browser's Print command doesn't work. There are three solutions:
-
For turning in homework, a class may allow you to submit the notebook file (
.ipynbfile) electronically (the graders will handle printing). -
The best way to print is currently to use IPython's nbconvert utility. For example, if you have a file
mynotebook.ipynb, you can runipython nbconvert mynotebook.ipynbto convert it to an HTML file that you can open and print in your web browser. This requires you to install IPython, Sphinx (which is automatically installed with the Anaconda Python/IPython distribution), and Pandoc on your computer. -
If you post your notebook in a Dropbox account or in some other web-accessible location, you can paste the URL into the online nbviewer to get a printable version.