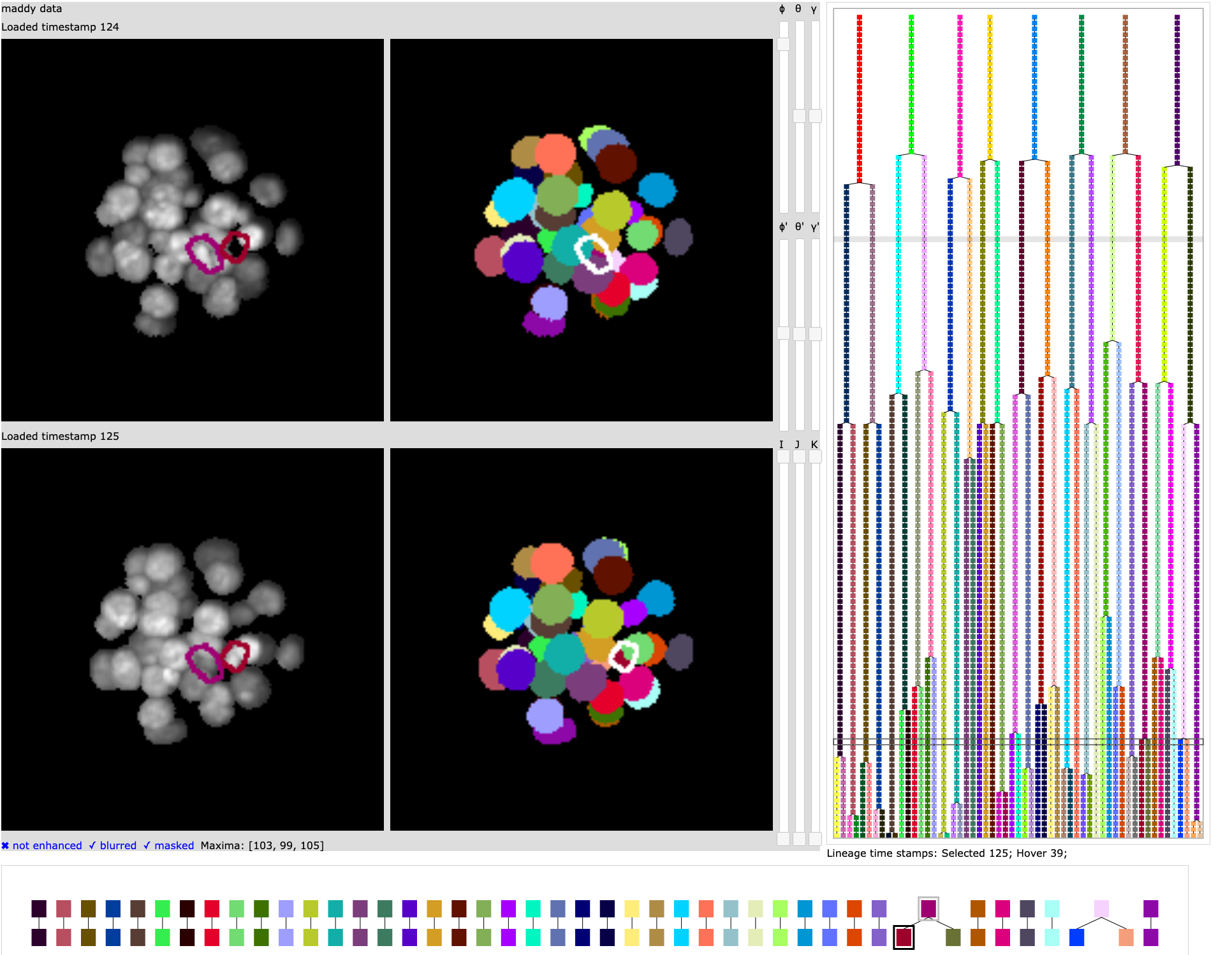
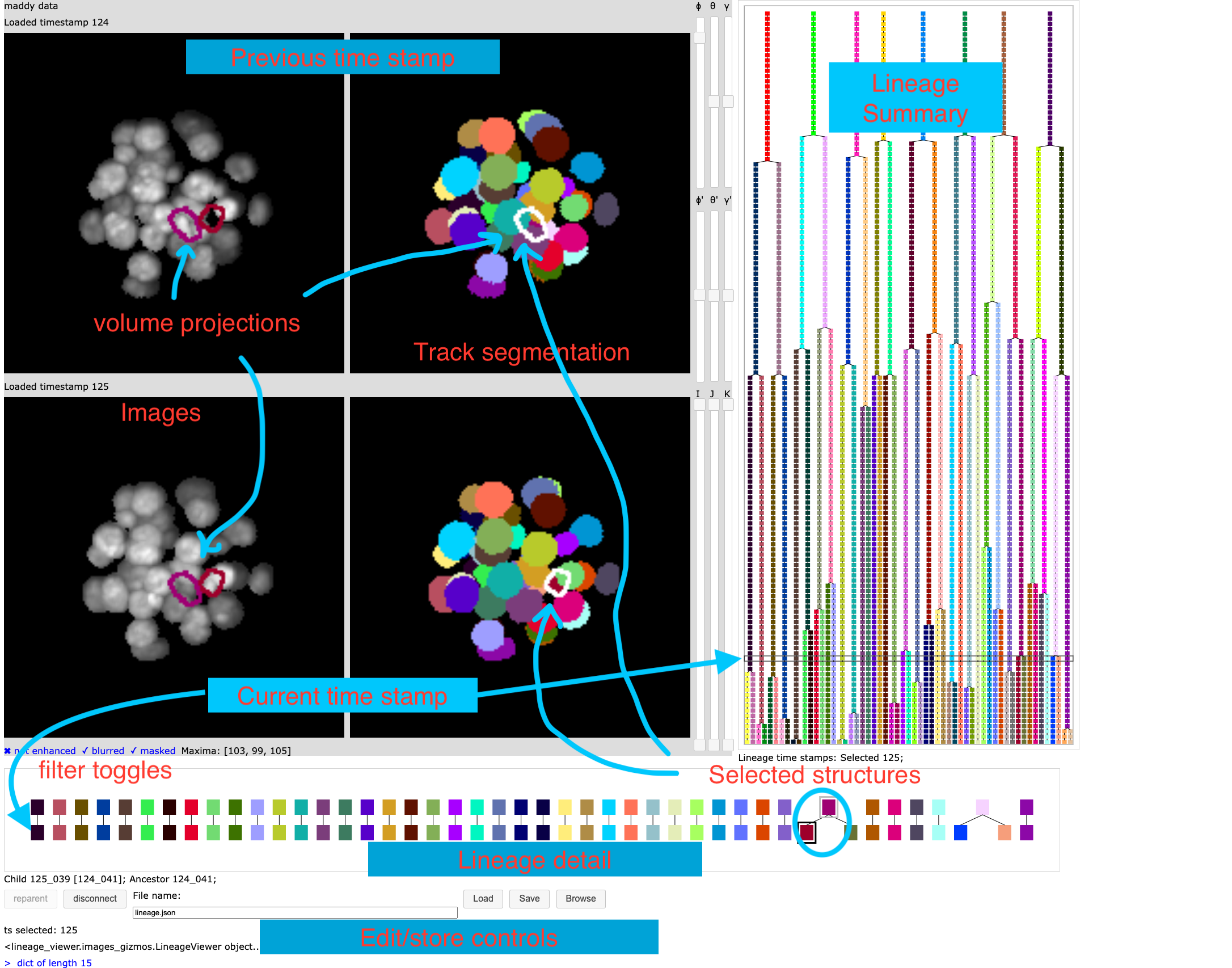
Graphical interface for editing a lineage and viewing related microscopy images.
This package implements a graphical interface designed for use in microscopy studies of early embryology. The visualization allows the user to explore the relationship between time sequences of microscopy image volumes of an embryo, segmentation volumes for the embryo identifying cell structures, and cell lineage trees for the cells of the embryo.
The interface includes the ability to modify, store, and load the lineage tree.
The interface was specifically designed for use in studies of early mouse embryology using light sheet microscopy extracted from a live embryo over many days.
This package requires Python 3.7 or higher. If you need to install an appropriate Python I recommend using conda installed in single user mode (for easy maintenance) http://bit.ly/tryconda.
The package install procedure is not fully automated.
Please install the following dependencies manually in the following order.
pip install git+https://github.com/AaronWatters/H5Gizmos
pip install git+https://github.com/AaronWatters/jp_doodle
pip install git+https://github.com/AaronWatters/array_gizmos
pip install git+https://github.com/flatironinstitute/mouse_embryo_labeller
Then clone this repository and in the top level folder of the repository install the module in development mode as follows:
git clone https://github.com/flatironinstitute/lineage_viewer.git
cd lineage_viewer
pip install -e .The repository includes a script with some data for an example lineage which should work "out of the box." The sample includes a full lineage tree description but only includes image data for timestamps 1 and 2 of the tree.
To try the example lineage go to the lineage_viewer/examples/lineage_sample
directory and run the load_sample_lineage.py script.
For example on my laptop:
(base) C02XD1KGJGH8:lineage_sample awatters$ cd ~/repos/lineage_viewer/
(base) C02XD1KGJGH8:lineage_viewer awatters$ cd examples/lineage_sample/
(base) C02XD1KGJGH8:lineage_sample awatters$ python load_sample_lineage.py
Open gizmo using link (control-click / open link)
<a href="http://127.0.0.1:56998/gizmo/http/MGR_1671120997917_6/index.html" target="_blank">Click to open</a> <br>
GIZMO_LINK: http://127.0.0.1:56998/gizmo/http/MGR_1671120997917_6/index.html
On my Mac laptop I have to confirm that Python can accept incoming connections in a pop up dialogue.
Then open the provided link URL in a browser to start the interface. When the interface loads click on timestamp 2 near the top of the lineage tree summary to load the images for timestamps 1 and 2 (as discussed in more detail below).
The Github repository for this package includes a number of example scripts and data files
in the examples folder:
examples/lineage_sample-- This folder includes a complete "stand alone" example data set.examples/lineage_sample/load_sample_lineage.py-- The "launch script" for the example data set which loads a JSON description of the lineage.examples/lineage_sample/load_labels_only.py-- A launch script which loads an "empty" lineage with nodes inferred from volume labels.examples/lineage_sample/LineageGraph.json-- The JSON dump file defining the lineage forest.examples/lineage_sample/nuclei_reg8_2.tif-- Microscopy volume for time stamp 2.examples/lineage_sample/nuclei_reg8_1.tif-- Microscopy volume for time stamp 1.examples/lineage_sample/label_reg8_1.tif-- Nucleus labelled volume for time stamp 1.examples/lineage_sample/label_reg8_2.tif-- Nucleus labelled volume for time stamp 2.examples/lineage_sample/get files.ipynb-- Historical notebook used to get the data filesexamples/Combined.json-- The JSON dump file for220827_stack1examples/nuc_to_cells.csv-- The membrane label correction CSV file for220827_stack1(from Masha).examples/load_rusty_test_data.py-- An example script for loading data on therustycluster for220827_stack1using KLB volume data.examples/load_maddy_data.py-- An example script for loading data on therustycluster for220827_stack6using TIFF volume data.examples/awatters_test_no_parents.py-- An example script that loads nodes from JSON with no parentage relationships.
The remainder of this document describes the components of the interactive user interface followed by a discussion of how to create a viewer and attach it to data sources.
For better performance do not launch the interface on rusty (or another compute cluster) from the login nodes.
Instead allocate a node using srun (or similar) to get a node with lower usage and better file system access:
(base) C02XD1KGJGH8:lineage_viewer awatters$ ssh rusty
No Slurm jobs found on node.
Last login: Tue Dec 13 09:03:14 2022 from 172.28.53.93
[awatters@rusty1 ~]$ srun -N1 --pty bash -i
(base) bash-4.4$ cd repos/lineage_viewer/examples/
(base) bash-4.4$ python load_maddy_data.py
....
Open gizmo using link (control-click / open link)
<a href="http://10.250.145.112:34875/gizmo/http/MGR_1671465177684_6/index.html" target="_blank">Click to open</a> <br>
GIZMO_LINK: http://10.250.145.112:34875/gizmo/http/MGR_1671465177684_6/index.html
....The graphical interface for the viewer displays in a web browser. For flexibility the example scripts do not start the browser interface automatically but instead provide a URL link so the user may choose what browser instance to use. For example
(base) bash-4.4$ python load_rusty_test_data.py
...
Open gizmo using link (control-click / open link)
<a href="http://10.128.146.57:59663/gizmo/http/MGR_1671116590930_6/index.html" target="_blank">Click to open</a> <br>
GIZMO_LINK: http://10.128.146.57:59663/gizmo/http/MGR_1671116590930_6/index.html The user may open the generated link http://10.128.146.57:59663/gizmo/http/MGR_1671116590930_6/index.html
by control-clicking the link (on a Mac OS for example). In this case the "back end" parent process runs
on the rusty cluster and the "front end" user interface runs on my Mac laptop.
The interface starts with no timestamp selected. With no timestamp selected only the lineage tree summary in the right panel displays.
Select a current time stamp by clicking on the lineage tree summary on the right. When a time stamp is selected the interface looks like the annotated image below.
This interactive interface presents the following components
- A time sequence of microscopy image stacks (volumes) of an embryo.
- A derived timesequence of labelled volumes identifying cell structures of the embryo. The structures can be cell nuclei, cell membrane volumes, or other structures of interest. The "shape" of the cell volumes should match the "shape" of the microscopy volumes.
- A derived cell lineage tree which identifies the parent/child relationship for the cells over the time sequence.
- The interface highlights a selected timestamp and the previous timestamp.
- The user can focus on a "selected structure" in the current and previous timestamp.
- The user can slice and rotate the images.
The interface is intended to allow the researcher to explore the data sets and possibly modify the lineage tree.
The following sections discuss the components of the interface in detail:
The lineage tree summary on the right side of the interface displays the lineage of parent/child relationships among cells for the timestamps, with earlier time stamps at the top. Cells with no parent or child relationship are not displayed. The user may focus on one of the timestamps and its immediate previous timestamp by clicking the canvas. The user can move the focus up or down using the arrow keys when the mouse is hovered over the canvas.
The interface highlights a current and previous timestamp indicated by a rectangle on the lineage tree. Initially there is no current timestamp and only the lineage tree summary displays.
When the current time stamp is selected the relationship between the cells of the current timestamp and the previous timestamp is displayed in the lineage detail at the bottom of the interface. All cells of both timestamps will display in the detail view even if they don't have parent/child relationships.
The user can identify a "selected" structure for the current and previous timestamps by clicking the rectangles that correspond to that structure. The selections will be highlighted by outer rectangles in the timestamp detail and their projections will appear in the images (as described below).
The images on the left side display maximal value projections of the source microscopy images. The volume for the current timestamp appears at the bottom and the volume for the previous timestamp appears at the top.
Outlines of selected cell structures are projected in the images even if they are obscured behind other structures.
Filter toggles below the microscopy images control whether the microscopy images are filtered. Click the controls to turn them on and off.
- The "mask" toggle enables masking of the images so that regions that are not labelled show as black background.
- The "blur" toggle controls whether background noise is "averaged out" and the dynamic range of values is stretched.
- The "enhance" control increases the image contrast.
The colored images to the right of the microscopy images show segmentation volumes extruded to 2 dimensions. The volume for the current timestamp appears at the bottom and the volume for the previous timestamp appears at the top. The labelled volumes are colored to match the corresponding entry in the lineage tree. Any labelled volumes that do not correspond to a node of the lineage tree show as gray.
Outlines of selected cell structures are projected in the image in white even if they are obscured behind other structures. The user can select a structure by clicking the corresponding colored region.
The microscopy and labelled volumes can be rotated using the rotation sliders to the right of the image area.
- Sliders "ϕ", "θ", "γ" apply rotations to all image volumes.
- Sliders "ϕ'", "θ'", "γ'" apply rotations to only the current timestamp volumes.
Slicing range sliders appear to the right of the images below the rotation sliders. These sliders apply slicing in the I, J, and K dimensions to the image volumes so the user can view structures in interior regions of the volume with outer structures sliced away.
The "reparent" and "disconnect" buttons at the bottom allow the user to modify the lineage tree structure by changing the parent relationship for the structure selected in the current timestamp.
If there is a structure selected in both the current and previous time stamp then the "reparent" button will make the earlier structure the parent of the later structure in the lineage tree.
If a selected structure in the current time stamp has a parent in the lineage then the "disconnect" button will remove its parent relationship and leave the structure with no parent.
The "Save" and "Load" buttons at the bottom save and load the lineage tree structure to/from the file path named in the adjacent "file name" input area. The "Browse" button opens a dialog which allows the user to browse the file system for files or folders.
This section describes how to load a lineage into the lineage viewer and how to specify the files containing the volume data corresponding to the timestamps of the lineage.
In Python a images_gizmos.LineageViewer object is created using a
lineage_forest.Forest container
# Create and populate a Forest
forest = lineage_viewer.lineage_forest.Forest()
# ... define the lineage and specify the volume file locations ...
viewer = images_gizmos.LineageViewer(forest, side=600, title="Lineage 28")The side parameter determines the size of a side of the image rectangles.
The title string appears at the top of the user interface.
To initialize a viewer you need to create a Forest object and
- Provide accessor functions which load label volume and image volume data as numpy arrays from the file system.
- Define the nodes of the lineage and identify the labels and timestamps associated with each node.
- Optionally define any parent/child relationship between the nodes.
The forest object encapsulates the lineage forest and
methods for finding the image and label volumes for a timestamp number (called an ordinal below).
The following code fragment from the examples/load_maddy_data.py script
illustrates two ways to define image loaders for a LineageViewer.
At the top the first F.use_trivial_null_loaders() method defines the loaders
to always return None indicating that the images for the timestamp ordinal
could not be found.
from mouse_embryo_labeller import tools
trivialize = False
if trivialize:
F.use_trivial_null_loaders()
else:
prefix = "/mnt/ceph/users/lbrown/MouseData/Maddy/220827_stack6/"
image_pattern = prefix + "registered_images/nuclei_reg8_%(ordinal)d.tif"
label_pattern = prefix + "registered_label_images/label_reg8_%(ordinal)d.tif"
def img_loader(ordinal, pattern=image_pattern):
subs = {"ordinal": ordinal} # files are zero based?
path = pattern % subs
print("attempting to load path", repr(path))
if os.path.exists(path):
return tools.load_tiff_array(path)
else:
return None # no data for this timeslice.
def label_loader(ordinal):
return img_loader(ordinal, label_pattern)
F.image_volume_loader = img_loader
F.label_volume_loader = label_loader
test_ordinal = 29
img = F.load_image_for_timestamp(test_ordinal)
assert img is not None, "Could not load image for: " + repr(test_ordinal)
lab = F.load_labels_for_timestamp(test_ordinal)
assert lab is not None, "Could not load image for: " + repr(test_ordinal)in the second method the img_loader function loads an image for a timestamp ordinal
from a TIFF file using a file pattern. In this case the label volume and image volume for timestamp 21
are stored in the files
/mnt/ceph/users/lbrown/MouseData/Maddy/220827_stack6/registered_images/nuclei_reg8_21.tif
/mnt/ceph/users/lbrown/MouseData/Maddy/220827_stack6/registered_label_images/label_reg8_21.tif
The lines
F.image_volume_loader = img_loader
F.label_volume_loader = label_loaderattach the loader functions to the forest object F.
The forest object also includes a convenience method for loading KLB format files.
To use this method you must install the pyklb Python package (which can be a bit tricky).
The following fragment from the examples/load_rusty_test_data.py script shows how to use the
forest.load_klb_using_file_patterns method:
label_pattern = "labels/klbOut_Cam_Long_%(ordinal)05d.crop_cp_masks.klb"
image_pattern = "images/klbOut_Cam_Long_%(ordinal)05d.crop.klb"
F.load_klb_using_file_patterns(
image_pattern=image_pattern,
label_pattern=label_pattern,
)The forest object
maintains a collection of Nodes and the parent/child relationship between the nodes
and provides other related computational methods. This package includes convenience
functions to automate constructing a forest and the forest object also includes
methods for adding nodes and parent relationships in other scripts.
The lineage_forest.make_forest_from_haydens_json_graph convenience function
populates a forest from a JSON data file structured similar to examples/Combined.json.
Such files are derived from upstream Matlab based processes.
The following code fragment from examples.load_rusty_test_data illustrates how
to construct a forest from Combined.json:
label_assignment = {}
... construction of assignment omitted ...
fn = "Combined.json"
json_ob = json.load(open(fn))
F = lineage_forest.make_forest_from_haydens_json_graph(json_ob, label_assignment=assignment)The label_assignment mapping describes how to map node names to numeric labels.
The assignment may be omitted (None) if the correct label can be derived from the node name.
For example the label number derived from the node name 009_004 is 4.
Please examine the source code for lineage_forest.make_forest_from_haydens_json_graph for
additional options and details about this function.
The examples/lineage_sample/load_labels_only.py prepares a lineage forest for the lineage_sample data
with no parent relationships. In this case the nodes for the timestamps are inferred from the
labels in the label volumes. The forest object F provides a convenience method to infer the labels,
used in this code fragment:
# **** Create nodes inferred from label volumes for timestamps 1 and 2
for timestamp_ordinal in range(1, 3):
F.create_nodes_for_labels_in_timestamp(timestamp_ordinal)The forest object provides methods for adding nodes to a forest and defining the
parentage relationship between nodes. These methods can be used in Python scripts to
construct lineage forests.
The
node = forest.add_node(node_id, ordinal, label=None)adds a node to a forest at a timestamp ordinal. The node_id must be a unique string.
the label number defines the label corresponding to this node in the label volume for
the timestamp number ordinal. If the node does not correspond to a label of the label volume
it may be omitted.
The parent_node.set_child(child_node) method of the Node object define a parent/child relationship
in the forest.
Please look to the source code of lineage_forest.make_forest_from_haydens_json_graph for example
uses of these methods.
It is possible to build the lineage forest structure manually using only the viewer interface by defining only node names for a forest but no parentage relationships.