HATCHet is an algorithm to infer allele and clone-specific copy-number aberrations (CNAs), clone proportions, and whole-genome duplications (WGD) for several tumor clones jointly from multiple bulk-tumor samples of the same patient or from a single bulk-tumor sample. HATCHet has been designed and developped by Simone Zaccaria in the group of prof. Ben Raphael at Princeton University. The full description of the algorithm, the comparison with previous state-of-the-art methods, and its application on published cancer datasets are described in
Simone Zaccaria and Ben Raphael, 2018
The novel simulationg framework, MASCoTE, is available at
The simulated data, the results of all the methods considered in the comparison, the results of HATCHet on the published whole-genome multi-sample tumor sequencing datasets ((Gundem et al., Nature, 2015) and (Makohon-Moore et al., Nature genetics, 2017)) and all the related analyses are available at
This repository includes a quick overview of the HATCHet's algorithm and software, detailed instructions for installation and requirements, a script to execute the full pipeline of HATCHet, demos to illustrate how to apply HATCHet on different datasets with different features, a list of current issues, and contacts.
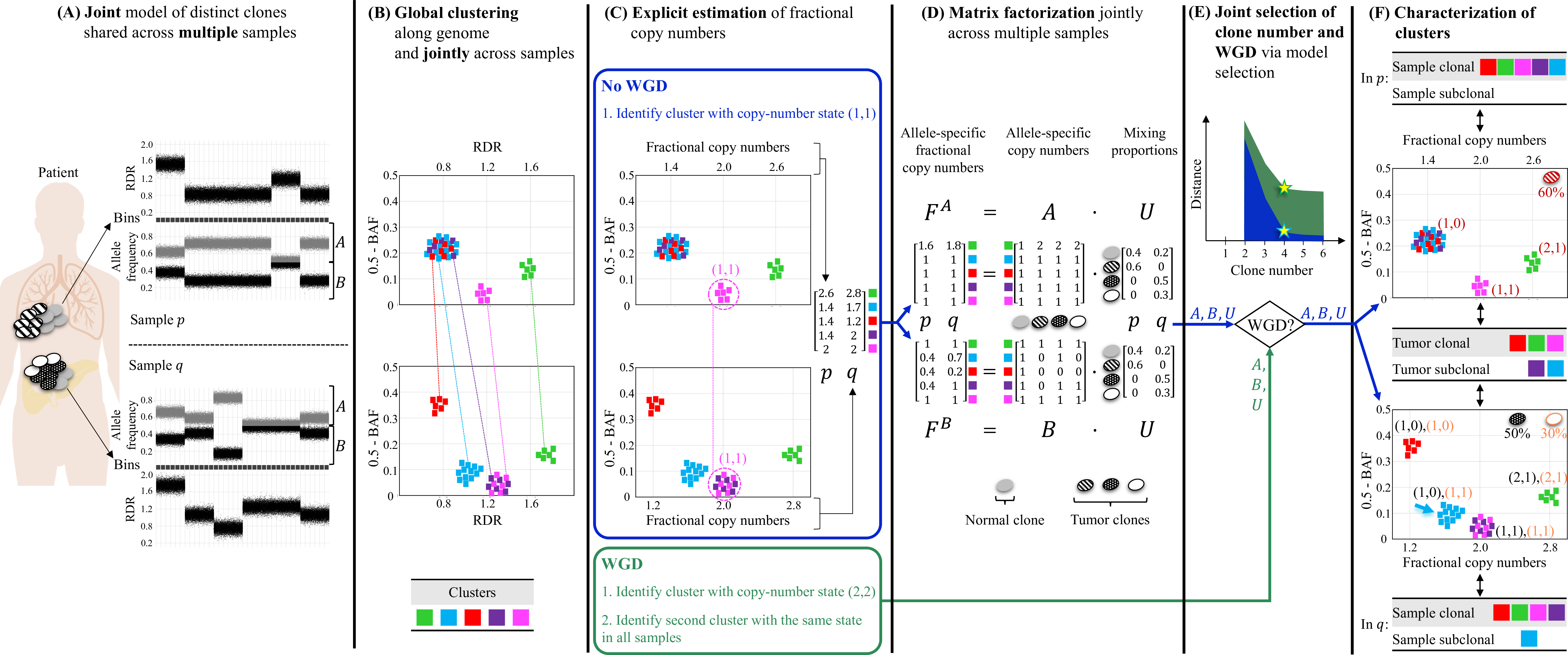
Overview of HATCHet algorithm. (A) HATCHet analyzes the read-depth ratio (RDR) and the B-allele frequency (BAF) in bins of the reference genome (black squares) jointly from multiple tumor samples. Here, we show two tumor samples p and q. (B) HATCHet globally clusters the bins based on RDR and BAF along the entire genome and jointly across samples p and q. Each cluster (color) includes bins with the same copy-number state within each clone present in p or q. (C) HATCHet estimates the fractional copy number of each cluster. If there is no WGD, the identification of the cluster (magenta) with copy-number state (1, 1) is sufficient and RDRs are scaled correspondingly. If a WGD occurs, HATCHet finds the cluster with copy-number state (2, 2) (same magenta cluster) and a second cluster having an identical copy-number state in all tumor clones. (D) HATCHet factorizes the allele-specific fractional copy numbers F^A, F^B into the allele-specific copy numbers A, B, respectively, and the clone proportions U. Here there is a normal clone and 3 tumor clones. (E) HATCHet's model selection criterion identifies the matrices A, B and U in the factorization while evaluating the fit according to both the inferred number of clones and presence/absence of a WGD. (F) Clusters are classified by their inferred copy-number states in each sample. Sample-clonal clusters have a unique copy-number state in the sample and correspond to evenly-spaced positions in the scaled RDR-BAF plot (vertical grid lines in each plot). Sample-subclonal clusters (e.g. cyan in p) have different copy-number states in a sample and thus correspond to intermediate positions in the scaled RDR-BAF plot. Tumor-clonal clusters have identical copy-number states in all tumor clones -- thus they are sample-clonal clusters in every sample and preserve their relative positions in scaled-RDR-BAF plots. In contrast, tumor-subclonal clusters have different copy-number states in different tumor clones and their relative positions in the scaled RDR-BAF plot varies across samples (e.g. purple cluster).
The current implementation of HATCHet is composed of two sets of modules:
(1) The core modules of HATCHet are designed to efficiently solve a challenging constrained and distance-based simultaneous matrix factorization which aim to infer allele and clone-specific copy numbers and clone proportins from fractional copy numbers. The module are implemented in C++11 and are included in src folder.
(2) The utility modules of HATCHet perform several different tasks that are needed to process the raw data, perform steps of the HATCHet's algorithm needed for the factorization, and process the results. These task include reading/calling germinal single-point mutations, counting reads from a BAM file, combining the read counts and other information, segmenting through HATCHet's global approach, plotting very useful information, etc. THese modules are implemented in python2.7 and are included in util and bin folders.
The setup process is composed of 4 steps:
- Dependencies: the installation of all the required dependecies; this is a one-time process.
- Compilation: the compilation of HATCHet; this is a one-time process.
- Using Gurobi: the requirements to run Gurobi; these need to be satisfied before every run.
- Required data: the requirements for considered data; these need to be satisfied whenever using new data.
The core module of HATCHet is written in C++11 and thus require a modern C++ compiler that is supporting this (GCC >= 4.8.1, or Clang). In addition, the core module has the following dependencies:
- CMake (>= 3.0), this is needed to drive the compilation. OPTIONAL: we suggest the use of CCMake to facilitate the specifications of other dependencies.
- Gurobi (>= 6.0), this is used becase the coordinate-method applied by HATCHet is based on several integer linear programming (ILP) formulations. Gurobi is a commercial ILP solver with two licensing options: (1) a single-host license where the license is tied to a single computer and (2) a network license for use in a compute cluster (using a license server in the cluster). Both options are freely and easily available for users in academia here.
The utility modules are written in python2. These modules have the following dependencies, which need to be installed once:
| Name and link | REQUIRED | Usage | Comments |
|---|---|---|---|
| subprocess and multiprocess | REQUIRED | Multiprocessing and threading | Standard python modules |
| SAMtools and BCFtools | REQUIRED | reading BAM files, read counting, allele counting, and SNP calling | CURRENT ISSUE: Unfortunately, HATCHet currently support only the following versions of these softwares: 1.5, 1.6, and 1.7 |
| BNPY-dev or BNPY | REQUIRED | Non-parametric Bayesian clustering algorithm | We highly reccomend to install BNPY by simply cloning the repository in a specific directory and verify the related dependencies (Section Installing *bnpy* in the installation instructions). We also suggest to install the additional C++ libraries to improve the performance (Section Optional: Fast C++ libraries for HMM inference in the installation instructions) |
| pandas, matplotlib, seaborn | OPTIONAL | Plotting | Required only for plotting modules. Seaborn has been tested with version 0.8.1. While all these packages can be easily installed using standard package managment tools (e.g. pip) for the default python version on your OS, we suggest the usage of python virtual enviroments created directly through python or (reccomended) using Anaconda. In addition to packages with better perfomance, these enviroments guarantee to keep the same version of all pacjkages without affecting the standard installation of python on your OS. An enviroment specific for HATCHet can be created with the suggested versions of all required packages; the enviroment needs to be activated before every run or explicitly included in the pipeline script. |
Compilation is required to be executed only once for the core modules of HATCHet, while all the other modules are ready to use after the installation of the required packages and dependecies. This repository includes an automatic compilation process which requires only 4 simple steps.
- Get Gurobi: user simply needs to unpack Gurobi (Linux version, no other step is required) or to install it (Mac version, standard installation is in
/Library/). - Build Gurobi: user only needs to build Gurobi when using newer GXX compilers (>= 5.0) on Linux or having linking issues by following these steps (assuming Gurobi's home is
/path/to/gurobiXXXwhereXXXis the 3-digit version andYYYYY64is equal to eitherlinux64ormac64when a subfolder is present present):$ cd /path/to/gurobiXXX/YYYYY64/src/build/ $ make $ cp libgurobi_c++.a ../../lib/ - Link HATCHet to Gurobi: substitute the following line at the beginning of
FindGUROBI.cmakewithset(GUROBI_HOME "" )to link HATCHet with Gurobi.set(GUROBI_HOME "/path/to/gurobiXXX" ) - Build HATCHet: execute the following commands from the root of HATCHet's repository.
$ mkdir build $ cd build/ $ ccmake .. $ make
NOTE: some users experienced a failure of compilation with an error message similar to undefined reference to symbol 'pthread_create@@GLIBC_2.2.5'. To solve this issue, simply substitute the following line in the CMakeLists.txt file:
set( CMAKE_CXX_FLAGS "${CMAKE_CXX_FLAGS} -std=c++11" )with the following:
set( CMAKE_CXX_FLAGS "${CMAKE_CXX_FLAGS} -std=c++11 -pthread" )by simply adding -pthread. After this, simply re-compile from scratch.
When the compilation process fail or when the enviroment has special requirements, the user can manually specify the required paths to Gurobi by following the detailed intructions.
Every run of HATCHet (especially, the hatchet step) needs to use Gurobi which requires a valid license pointed by the enviromental variable GRB_LICENSE_FILE. This can be easily obtained depending on the type of free academic license available:
- Individual license. This license can be obtained easily by any academic user with an istitutional email. This license is user and machine-specific, meaning that the user needs to require a different license for every used machine. Assuming the license is stored at
/path/to/gurobi.lic, the user can easily use it by the following command:export GRB_LICENSE_FILE="/path/to/gurobi.lic"
- Multi-use license. This license can be used by multiple users on any machine in a cluster. This license can be obtained but needs to be requested by the IT staff of the user's institution. This license is tipically used in a machine cluster and only requires the following command (where the name of gurobi module can slightly change):
module load gurobi
As a valid license needs to be available for every run, user can simply add the required command to the pipeline script used to run HATCHet, instead of executing it before every new access to a machine.
HATCHet requires 3 input data:
- One or more BAM files containing DNA sequencing reads obtained from tumor samples of a single patient. Every BAM file contains the sequencing reads from a sample ad needs to be indexed and sorted. For example, each BAM file can be easily indexed and sorted using SAMtools. In addition, one can improve the quality of the data by processing the BAM files according to the GATK Best Practices.
- A BAM file containg DNA sequecing reads obtained from a matched-normal sample of the same patient of the considered tumor samples. The BAM file needs to be indexed and sorted as the tumor BAM files. Also, the BAM files can be processed as the tumor BAM files.
3. A human reference genome. Ideally, one should consider the same human reference genome used to align the sequencing reads in the given BAM files. The most-used human reference genome are available at GRC or UCSC. Observe that human reference genomes use two different notations for chromosomes: either
1, 2, 3, 4, 5 ...orchr1, chr2, chr3, chr4, chr5 .... One needs to make sure all BAM files and reference genome share that same chromosome notation. When this is not the case, one needs to change the reference to guarantee consistency and needs to re-index the new reference (e.g. using SAMtools). Also, HATCHet requires that the name of each chromosome is the first word in each ID such that>1 [ANYTHING] ... \n>2 [ANYTHING] ... \n>3 [ANYTHING] ...or>chr1 [ANYTHING] ... \n>chr2 [ANYTHING] ... \n>chr3 [ANYTHING].
The repository includes all the components that are required to cover every step of the entire HATCHet's pipeline, starting from the processing of raw data reported in a BAM file through the analysis of the final results. We provide a script representing the full pipeline of HATCHet and we describe in details the whole script through a tutorial with instructions for usage. In addition we provide some demos which correspond to guided executions of HATCHet on some examples and explain in detail the usage of HATCHet when considering standard datasets, real datasets with high noise, and different kind of data. The repository also includes custom pipelines which adapts the full HATCHet's pipeline to special condition or integrates pre-processed data belonging to different pipelines. Moreover, the implementation of HATCHet is highly modular and one can replace any HATCHet's module with any other method to obtain the required results (especially for the pre-processing modules). As such, we also provide here an overview of the entire pipeline and we describe the details of each step in a dedicated section of the manual. Last, we provide reccomendations, especially for noisy datasets or with deifferent features, to guide the user in the interpration of HATCHet's inference, explain how to perform quality control to guarantee the best-quality results, and describe how the user can control and tune some of the parameters to obtain the best-fitting results.
We provide an exemplary BASH script that implements the entire pipeline of HATCHet. This script and its usage are described in detailed in a guided tutorial. The user can simply use the script for every execution of HATCHet on different data by copying the script inside the running directory and changing the corresponding paths of the required data and dependecies at the beginning of the script, as described in the guided tutorial.
Each demo is an exemplary and guided execution of HATCHet on a dataset included in the corresponding demo's folder of this repository (inside examples). The demos are meant to illustrate how the user should apply HATCHet on different datasets characterized by different features, noise, and kind of data. In fact, the default parameters of HATCHet allow to succesfully analyze most of the datasets but some of these may be characterized by special features or higher-than-expected variance of the data. Understanding the functioning of HATCHet, assessing the quality of the results, and tuning the few parameters needed to fit the unique features of the considered data thus become crucial to guarantee to always obtain the best-quality results. These are the goals of these demos. More specifically, each demo is simultaneously a guided description of the entire example and a BASH script, which can be directly executed to run the complete demo after setting the few required paths at the beginning of the file. As such, the user can both read the guided description as a web page and run the same script to execute the demo. At this time the following demos are available (more demos will be added in the near future):
| Name | Demo | Description |
|---|---|---|
| demo-WGS-sim | demo-wgs-sim | A demo on a typical WGS (whole-genome sequencing) multi-sample dataset with standard noise and variance of the data |
| demo-WGS-cancer | demo-wgs-cancer | A demo on a cancer WGS (whole-genome sequencing) multi-sample dataset with high noise and variance of the data |
| demo-WES | demo-wes | A demo on a cancer WES (whole-exome sequencing) multi-sample dataset, which is typycally characterized by very high variance of RDR |
The repository includes custom pipelines which have been designed to adapt the complete pipeline of HATCHet to special conditions or to integrate the processed data produced by other pipelines. Each custom pipeline is a variation of the main HATCHet's pipeline, we thus recommend the user to always first carefully understand the main BASH script through the corresponding guided tutorial and to carefully understand the provided demos to properly apply HATCHet for best-quality results. Each custom pipeline also includes a specific demo which represent a guided and executable example on example data.
| Name | Description | Script | Demo | Variations |
|---|---|---|---|---|
| GATK4-CNV | Custom pipeline for segmented files from GATK4 CNV pipeline | custom-gatk4-cnv.sh | demo-gatk4-cnv.sh | This custom pipeline takes the input the segmented files which already contain the estimated RDR and BAF. As such, the first pre-processing steps of HATCHet (binBAM, deBAF, and comBBo) are not needed; for this reason, the following depdencies SAMtools and BCFtools and the following required data, human reference genome, matched-normal sample, and BAM files, are not needed in this case. |
The full pipeline of HATCHet is composed of 7 sequential steps, starting from the required input data. The descrition of each step also includes the details of the corresponding input/output that are especially useful when one wants to replace or change some of the steps in the pipeline while guaranteeing the correct functioning of HATCHet.
| Order | Step | Description |
|---|---|---|
| (1) | binBAM | This step splits the human reference genome into bins, i.e. fixed-size small genomic regions, and computes the number of sequencing reads aligned to each bin from every given tumor samples and from the matched normal sample. |
| (2) | deBAF | This step calls heterozygous germline SNPs from the matched-normal sample and counts the number of reads covering both the alleles of each identified heterozgyous SNP in every tumor sample. |
| (3) | comBBo | This step combines the read counts and the allele counts for the identified germline SNPs to compute the read-depth ratio (RDR) and B-allele frequency (BAF) of every genomic bin. |
| (4) | cluBB | This step globally clusters genomic bins along the entire genome and jointly across tumor samples, and estimate the corresponding values of RDR and BAF for every cluster in every sample. |
| (5) | BBot | This step produces informative plots concerning the computed RDRs, BAFs, and clusters. The information produced by this step are important to validate the compute clusters of genomic regions. |
| (6) | hatchet | This step computes allele-specific fractional copy numbers, solves a constrained distance-based simultaneous factorization to compute allele and clone-specific copy numbers and clone proportions, and deploys a model-selection criterion select the number of clone by explicitly considering the trade-off between subclonal copy-number aberrations and whole-genome duplication. |
| (7) | BBeval | This step analyzes the inferred copy-number states and clone proportions and produces informative plots jointly considering all samples from the same patient. In addition, this step can also combine results obtained for different patients and perform integrative analysis. |
All the components of HATCHet's pipeline use some basic parameters that allow to deal with data characterized by different features. The default values of these parameters allow one to succesfully apply HATCHet on most datasets. However, special or noisy datasets may require to tune some parameters. The user can deal with these cases by following the reccomendations reported here, reading the descriptions of the various steps, and using the informative plots to verify the results. In the following guides and recommentations, we guide the user in the interpration of HATCHet's inference, we explain how to perform quality control to guarantee the best-quality results, and we describe how the user can control and tune some of the parameters to obtain the best-fitting results. We thus split the reccomendations into distinct topics with dedicated descriptions.
| Reccomendation | Description |
|---|---|
| Analyze HATCHet inference | Interpret HATCHet's inference, quality and error control, and investigate alternative solutions. |
| Analyze global clustering | Interprent global clustering, quality and error control, and parameter tuning |
| Analyze different type of data | Tuning parameters to better analyzing different type of data as those from WES |
| Improve running time | Tips for improving running time of the whole pipeline |
HATCHet is in active development, please report any issue or question as this could help the devolment and imporvement of HATCHet. Current known issues with current version are reported here below:
- HATCHet currently only supports versions 1.5 -- 1.6 -- 1.7 of SAMtools and BCFtools
HATCHet currently only supports dev-bitbucket version of BNPY- The allele-swapping feature of comBBo has been temporarily disabled due to conflicts with recent SAMtools versions
- HATCHet has not been tested on Windows OS yet
A list of the major recent updates:
- HATCHet accepts now any reference genome (including non humans)
- HATCHet now supports both the
devand new GitHub version ofBNPY - HATCHet now handles genomic regions properly, reccomended for WES data
HATCHet's repository is actively mantained by Simone Zaccaria, currently a postodoctoral research associate at Princeton University in the research group of prof. Ben Raphael.