Electrocardiogram based electrolyte concentration prediction
Codebase for the paper "ECG-Based Electrolyte Prediction: Evaluating Regression and Probabilistic Methods".
Overview
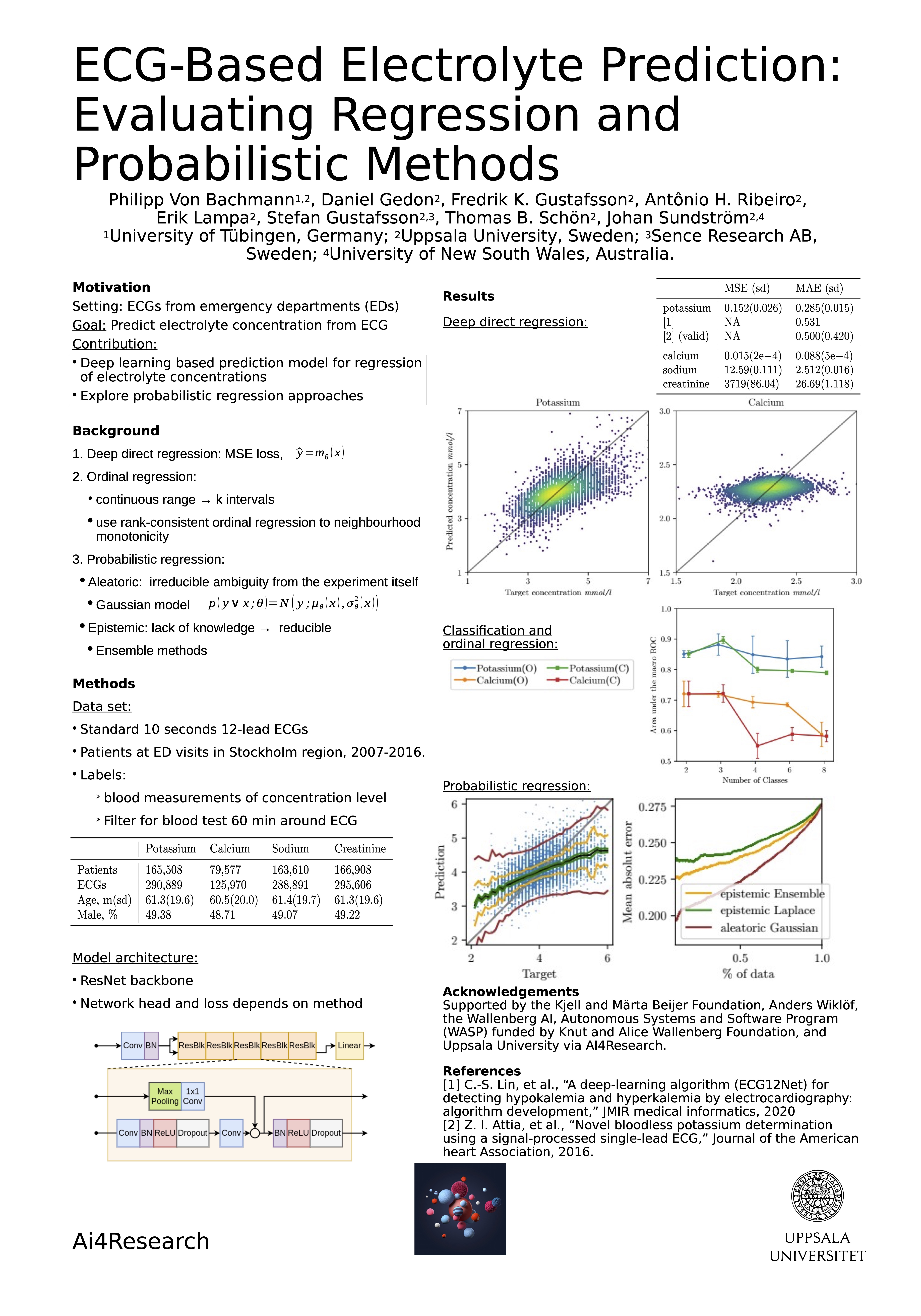
Here is a quick overview of the paper
Dataset
Unfortunately, our dataset is not publicly available. However you can implement your own dataset. In order to do this, please overwrite the method load_dset_electrolyte here. You can overwrite the whole method or alternatively change
- Loading of the ECG traces in line 62 - 66 (traces are given as
datapoints x leads x samples) - Loading of the metadata in line 61, which needs to contain
- A column whose name is given by the input argument
el_col, which contains the electrolyte concentration. - A column named "split", specifying the split of the individual datapoints.
- Additionally, columns named "ecg_age" and "is_male", to enable the use of this metadata, or alternatively disable these functionalities.
- A column whose name is given by the input argument
Training and Evaluating
A minimal example to train a model is
python experiments/training/train.py --folder OUTPUT_FOLDER --m MODEL_CONFIG --d DSET_CONFIG
which trains a regression model. MODEL_CONFIG is a json file which will be passed to the model constructor, an example can be found here, otherwise look in this file for the model constructors and their respective arguments. DSET_CONFIG is a json file as well, which specifies arguments passed to your modified dataset constructor.
The behaviour can be furter customized by adding specifying more arguments. The main ones are:
--taskchanges on which task to train the model, options are "regression", "classification", "ordinal", "gaussian".--bucketsif task is "classification" or "ordinal", uses these intervals to discretize the targets, should be passed as list of floats.--epochschange the number of epochs.
All additional arguments with descriptions can be retrieved by executing
python experiments/training/train.py --help
In order to evaluate a model, run
python experiments/evaluation/forward.py --model PATH_TO_MODEL --splits LIST_OF_SPLITS
where the LIST_OF_SPLITS are the dataset splits you defined when overwriting the method load_dset_electrolyte. This creates a csv file containing the predictions and targets which can then be evaluated.
Pretrained models
We release our pretrained models here which can be used to try on your own models. Each model folder contains the state_dict of the model, for a tutorial see Loading and saving models in PyTorch, as well as the config used during training.