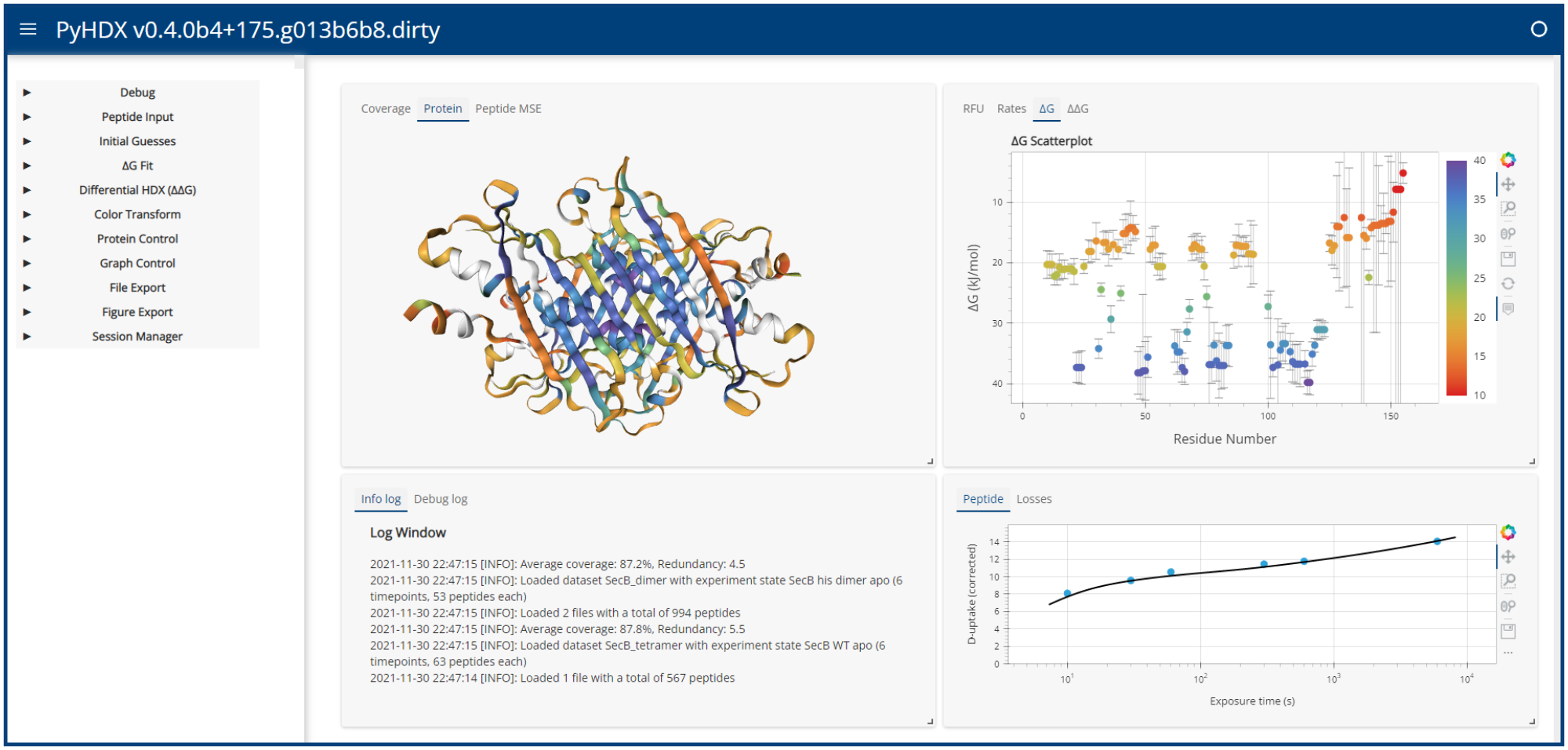
PyHDX is python project which can be used to derive Gibbs free energy from HDX-MS data. Currently version 0.3.2 is the stable release. Version 0.4.0 is released as beta.
Installation of the latest stable beta with `pip`:
pip install pyhdx==0.4.0b8Installation with web interface extra:
pip install pyhdx==0.4.0b8[web]Most up-to-date code examples are in the directory pyhdx/templates
To run the web server:
pyhdx servePlease refer to the docs for more details on how to run PyHDX.
The PyHDX web application is currently hosted at: http://pyhdx.jhsmit.org/main
A test file can be downloaded from here and here. (right click, save as).
The 0.4.0b4 version of PyHDX (featuring batch fitting / multiple states) is hosted at: http://pyhdx-beta.jhsmit.org/main
The latest beta docs are found here
Our Analytical Chemistry Publication describing PyHDX can be found here: https://doi.org/10.1021/acs.analchem.1c02155
The latest version (v2) of our biorxiv paper: https://doi.org/10.1101/2020.09.30.320887
Python code for analysis and generation the figures in the paper are here: https://github.com/Jhsmit/PyHDX-paper