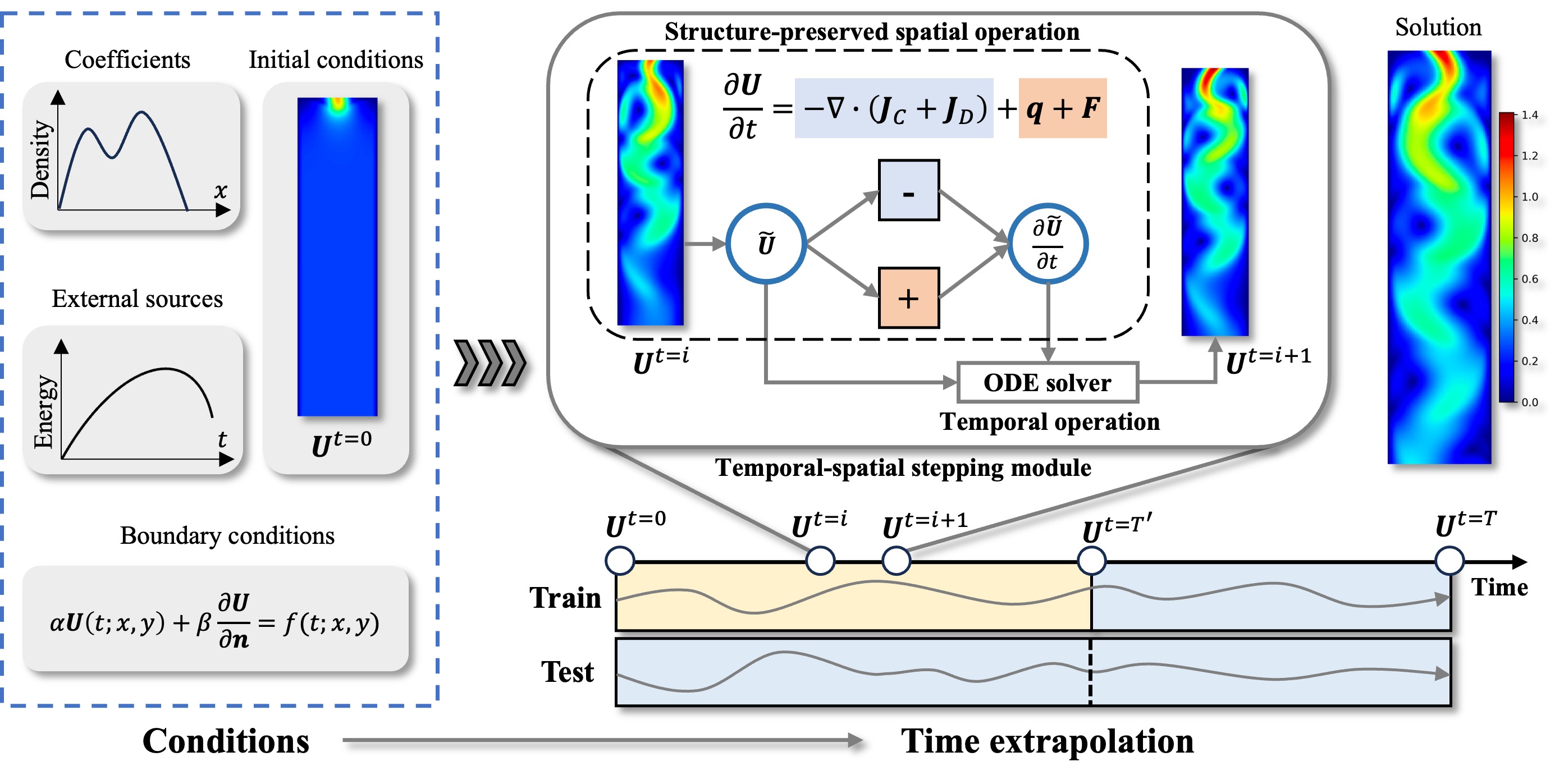
In the context of proxy modeling for process systems, traditional data-driven deep learning approaches frequently encounter significant challenges, such as substantial training costs induced by large amounts of data, and limited generalization capabilities. As a promising alternative, physics-aware models incorporate partial physics knowledge to ameliorate these challenges. Although demonstrating efficacy, they fall short in terms of exploration depth and universality. To address these shortcomings, we introduce a physics-aware proxy model (PAPM) that fully incorporates partial prior physics of process systems, which includes multiple input conditions and the general form of conservation relations, resulting in better out-of-sample generalization. Additionally, PAPM contains a holistic temporal-spatial stepping module for flexible adaptation across various process systems. Through systematic comparisons with state-of-the-art pure data-driven and physics-aware models across five two-dimensional benchmarks in nine generalization tasks, PAPM notably achieves an average performance improvement of 6.7%, while requiring fewer FLOPs, and just 1% of the parameters compared to the prior leading method.
The core contributions of this work are:
- The proposal of PAPM, a versatile physics-aware architecture design that fully incorporates partial prior knowledge such as multiple conditions, and the general form of conservation relations. This design proves to be superior in both training efficiency and out-of-sample generalizability.
- The introduction of a holistic temporal-spatial stepping module (TSSM) for flexible adaptation across various process systems. It aligns with the distinct equation characteristics of different process systems by employing stepping schemes via temporal and spatial operations, whether in physical or spectral space.
- A systematic evaluation of state-of-the-art pure data-driven models alongside physics-aware models, spanning five two-dimensional non-trivial benchmarks. Notably, PAPM achieved an average absolute performance boost of 6.7% with fewer FLOPs and only 1%-10% of the parameters compared to alternative methods.
As shown in following Table, we employ five datasets spanning diverse domains, such as fluid dynamics and heat conduction, detailed in Appendix. These datasets are categorized based on the Temporal-Spatial Stepping Method used in PAPM to highlight distinct equation characteristics in various process systems.
| Dataset | Category | Initial conditions | Boundary conditions | Coefficients | External sources |
|---|---|---|---|---|---|
| Burgers2d | Localized | ✔️ | Periodic | ✔️ | unknown |
| RD2d | Localized | ✔️ | No-flow Neumann | unknown | |
| NS2d | Spectral | ✔️ | Periodic | unknown | |
| Lid2d | Hybrid | Dirichlet, Neumann | ✔️ | ||
| NSM2d | Hybrid | ✔️ | Dirichlet, Neumann | ✔️ | unknown |
-
Localized Category.
-
Burgers2d [1] is a 2D benchmark PDE with periodic BC given by the equation
$\frac{\partial \boldsymbol{u}}{\partial t}=-\boldsymbol{u} \cdot \nabla \boldsymbol{u}+ \nu \Delta \boldsymbol{u}+\boldsymbol{f}$ . Here,$\boldsymbol{u}=(u_x, u_y)$ represents velocity. The aim is to predict subsequent frames of$\boldsymbol{u}$ under various initial conditions and viscosity$nu$ using the initial frames, while the forcing term$f$ remains unknown. -
RD2d [2] corresponds to a 2D F-N reaction-diffusion equation with no-flow Neumann BC,
$\frac{\partial \boldsymbol{u}}{\partial t}= \nu \Delta \boldsymbol{u}+\boldsymbol{f}$ . Here,$\boldsymbol{u}=(u, v)$ are the activator and inhibitor, respectively. The goal is to project subsequent frames under diverse initial conditions from the initial frames, with the source term as unknown.
-
Burgers2d [1] is a 2D benchmark PDE with periodic BC given by the equation
-
Spectral Category.
-
NS2d [3] is a dataset for incompressible fluid dynamics in vorticity form with periodic BC. The equation is
$\frac{\partial w}{\partial t}= -u\nabla w + \nu \Delta w + f$ , where$u$ is velocity,$w$ is vorticity, and$f$ is an unknown forcing term. The objective is to predict final frames from the initial frames of vorticity$w$ under varied initial conditions.
-
NS2d [3] is a dataset for incompressible fluid dynamics in vorticity form with periodic BC. The equation is
-
Hybrid Category.
-
Lid2d is a classical 2D dataset for incompressible lid-driven cavity flow with multiple BCs,
$\frac{\partial \boldsymbol{u}}{\partial t}=-\boldsymbol{u} \cdot \nabla \boldsymbol{u}+ \nu \Delta \boldsymbol{u} - \nabla p$ . The goal is to predict subsequent frames$(u,v,p)$ based on initial ones at differing viscosity$u$ , assuming only two flows are known. -
NSM2d is incompressible fluid dynamics with a magnetic field, described by
$\frac{\partial \boldsymbol{u}}{\partial t}=-\boldsymbol{u} \cdot \nabla \boldsymbol{u}+ \nu \Delta \boldsymbol{u} - \nabla p + F$ . The target is to predict subsequent frames of$(u,v,p)$ using the initial frames, with the forcing term$F$ as an unknown.
-
Lid2d is a classical 2D dataset for incompressible lid-driven cavity flow with multiple BCs,
Dataset Link:
- Burger2d (from PDENet++'s experiments): Burger2d
- RD2d (from PDEBench's experiments): RD2d
- NS2d (from FNO's experiments): NS2d
Add dataset folder if it does not exist, mdkir datasets, cd datasets. To store training models, you can build the folder:mkdir weights. Then, move datasets to this folder. You might need to modify the args and utils/load_data functions for your custom datasets.
Data Format:
The format of our dataset (Papm_Dataset) should be as follows:
Dataset = [
[Timestep1, X1, Y1],
[Timestep2, X2, Y2],
...
]
Physics_Parameters = [
[Param1],
[Param2],
...
]-
Timestep: (N_steps) numpy array, time steps for this sample - N_theta: dimension of time steps for this dataset
-
X,Y: (X_dim x Y_dim) numpy array, representing input mesh points
-
X_dim, Y_dim: input dimension of geometry
-
Physics_Parameters: (N_steps x 1) numpy array, representing physics parameter the model need,i.e,
Reynolds number -
Note: - For a single dataset sample, The number of points must match, i.e,
X.shape[0]=Y.shape[0], but it can vary with different dataset. - For physics parameters, the dimension must be the same as time_steps
To train PAPM model, parameters could be updated using argparser:
python main.py --file_link='./datasets/burgers_250_101_2_64_64.h5' --shuffle=True --model='papm' --gpu='cuda:0' --dataset='burgers' --train_bs=4 --val_bs=2 --seed=2024 --epoches=500 --test_accumulative_error=Truemain.py:main training functionutils:includes dataset, loss functionsmodels:folder for baselines and PAPM
- [1] Huang X, Li Z, Liu H, et al. Learning to simulate partially known spatio-temporal dynamics with trainable difference operators. arXiv preprint arXiv:2307.14395, 2023.
- [2] Takamoto M, Praditia T, Leiteritz R, et al. PDEBench: An extensive benchmark for scientific machine learning. Advances in Neural Information Processing Systems, 2022, 35: 1596-1611.
- [3] Li Z, Kovachki N, Azizzadenesheli K, et al. Fourier neural operator for parametric partial differential equations. arXiv preprint arXiv:2010.08895, 2020.