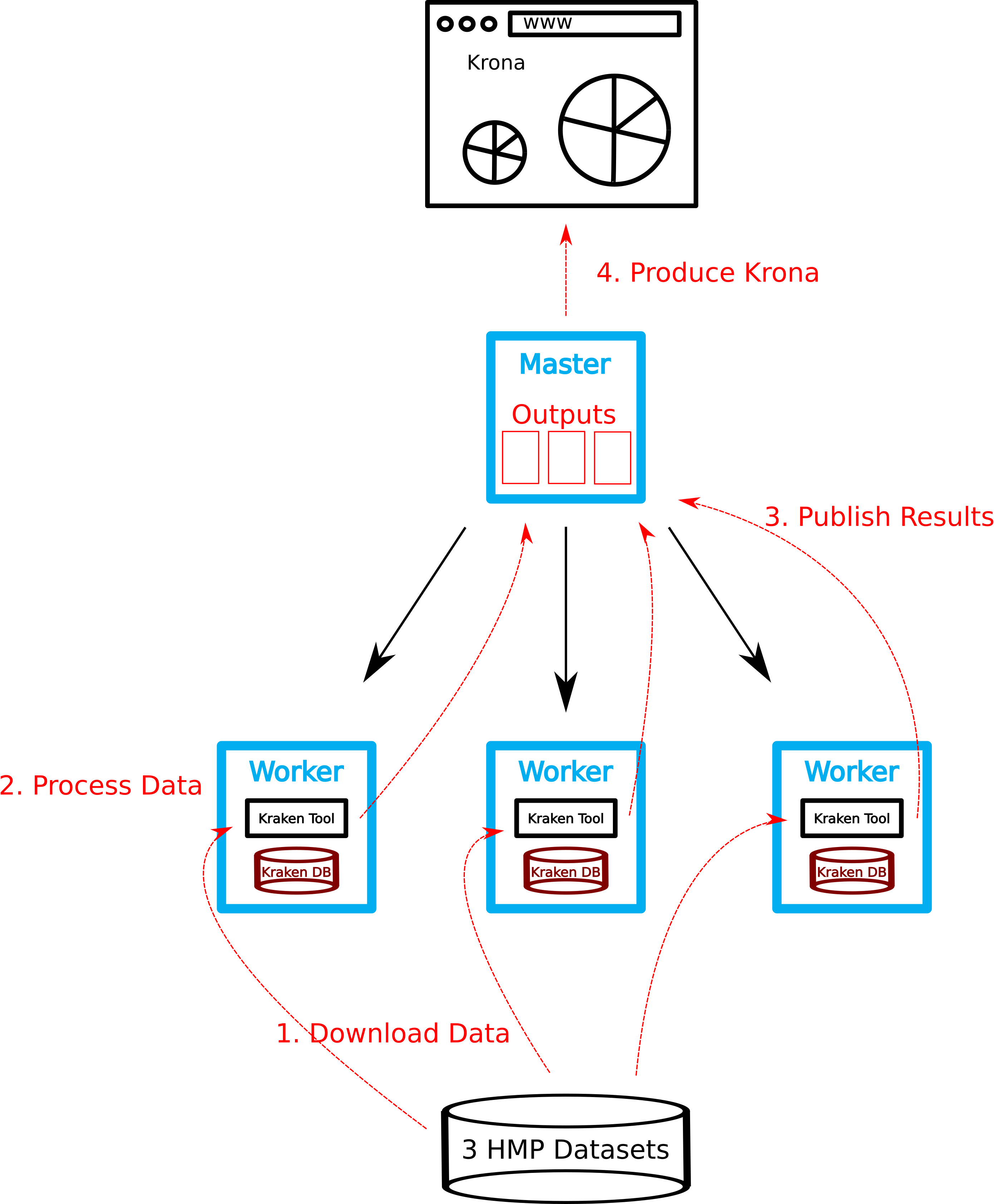
We want to use Kraken to analyze some FASTQ files from the Human Microbiome Project and use Krona to visualize the results. To makes the life easier we run ready-to-use dockerized versions of Kraken and Krona.
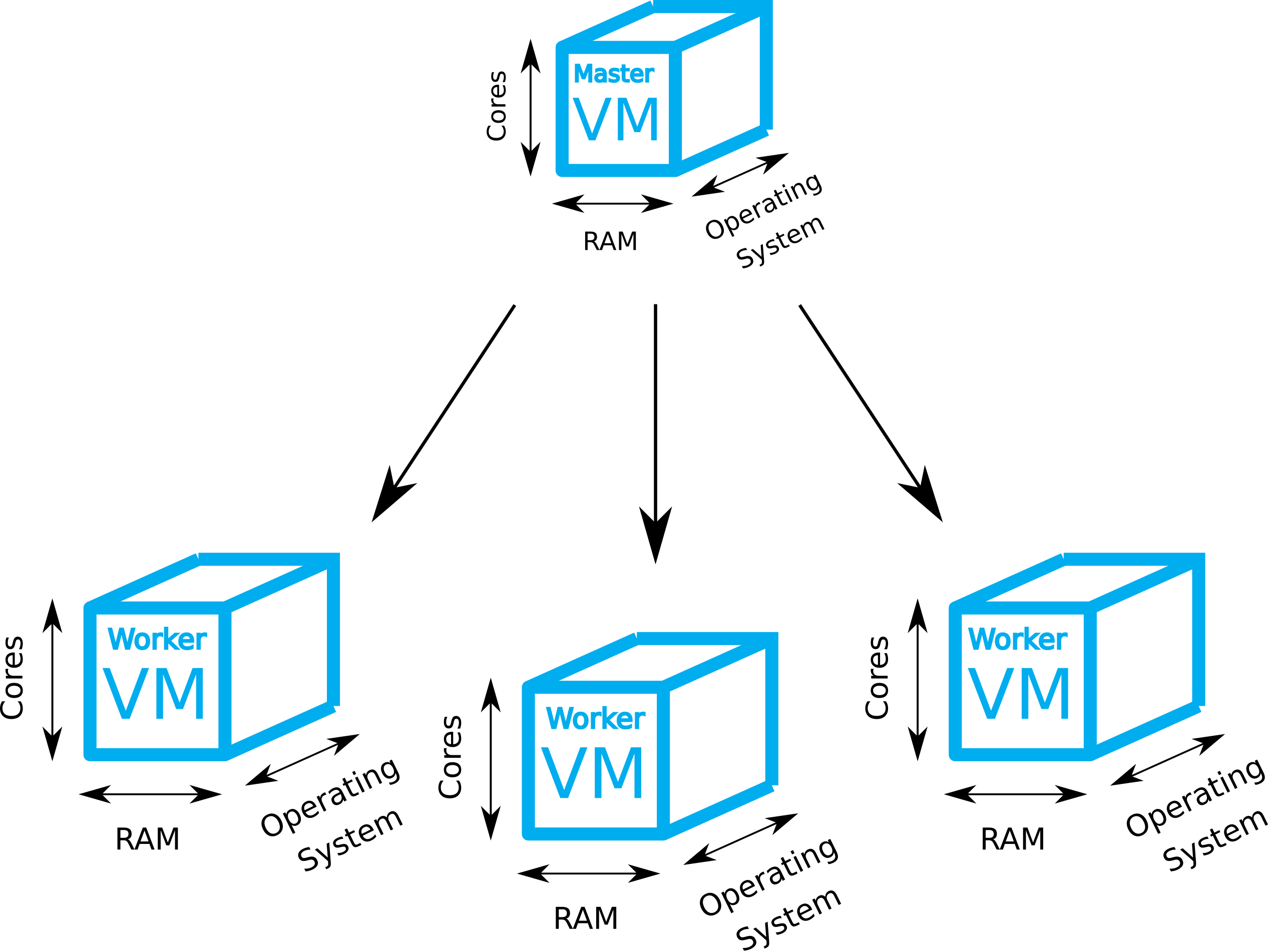
We assume that a bibigrid cluster ( master - 8 cores + 4 worker nodes - 16 cores each) with a configured GridEngine (GE) is already running. Please fetch this repository with
cd ~/
git clone https://github.com/pbelmann/kraken-demo.gitHint: The cloud users homedir is not shared between master and host. Any outputs from GE jobs are stored in users homedir as default. It is often a good idea to change this default behavior. We could change the environment setting of the GridEngine or just use the -cwd (Currrent Working Directory) argument and change into a shared fs before
cd /vol/spool/log
qsub -cwd ...
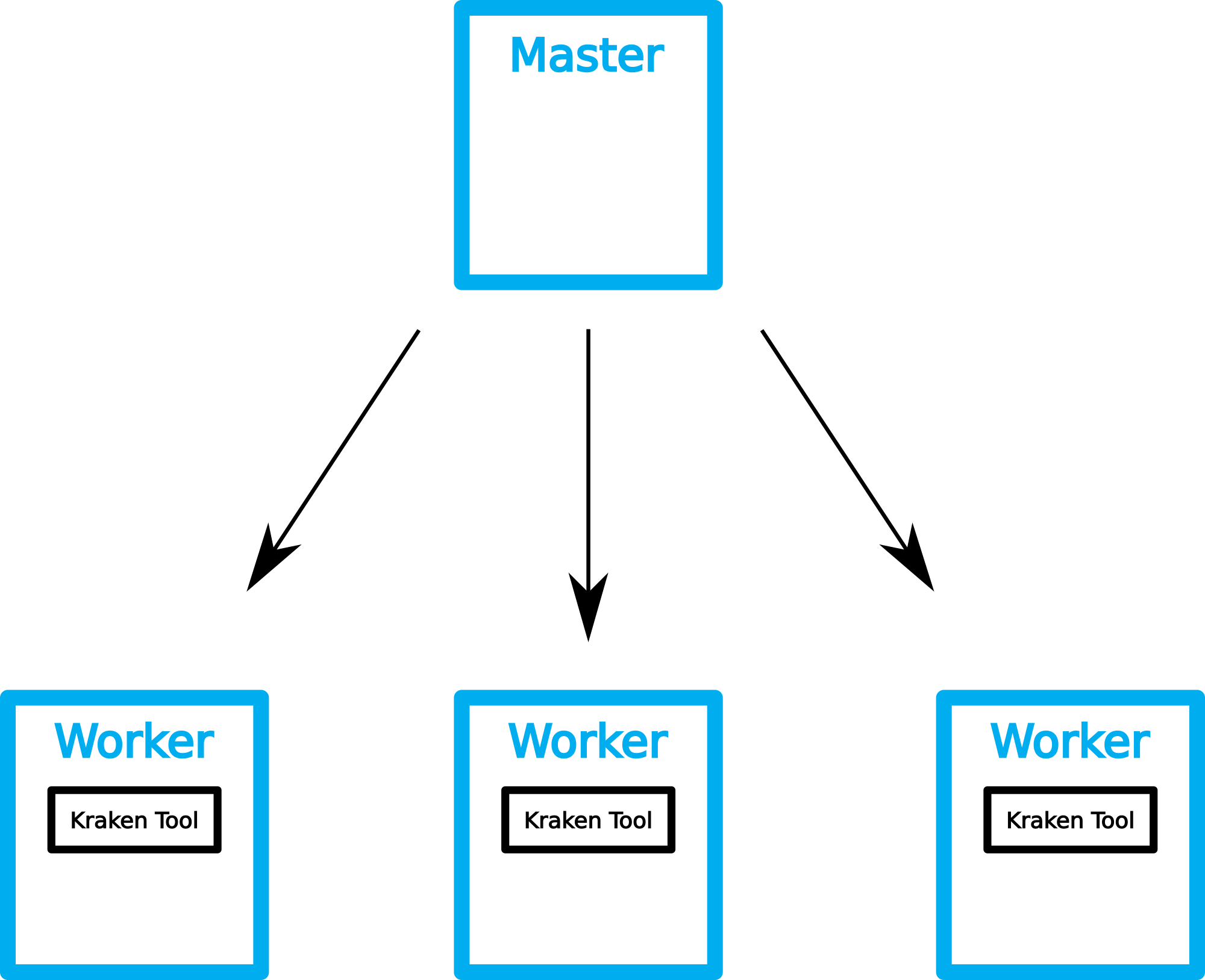
BioContainers is an open source container framework for bioinformatic software. We search BioContainers registry for a suitable Kraken container and write a small shell script that downloads the container once on each compute host. We can use the GE to distribute the jobs on the
cluster. The -pe option ensures, that we call the script only once on each host.
qsub -t 1-4 -pe multislot 16 -b y docker pull quay.io/biocontainers/kraken:1.0--pl5.22.0_0
This step seems to be unneccessary, because docker pulls a container if it isn't locally available. However, separate this step from rest of pipeline can speed up a analysis in the case you run more than one job in parallel on one host.
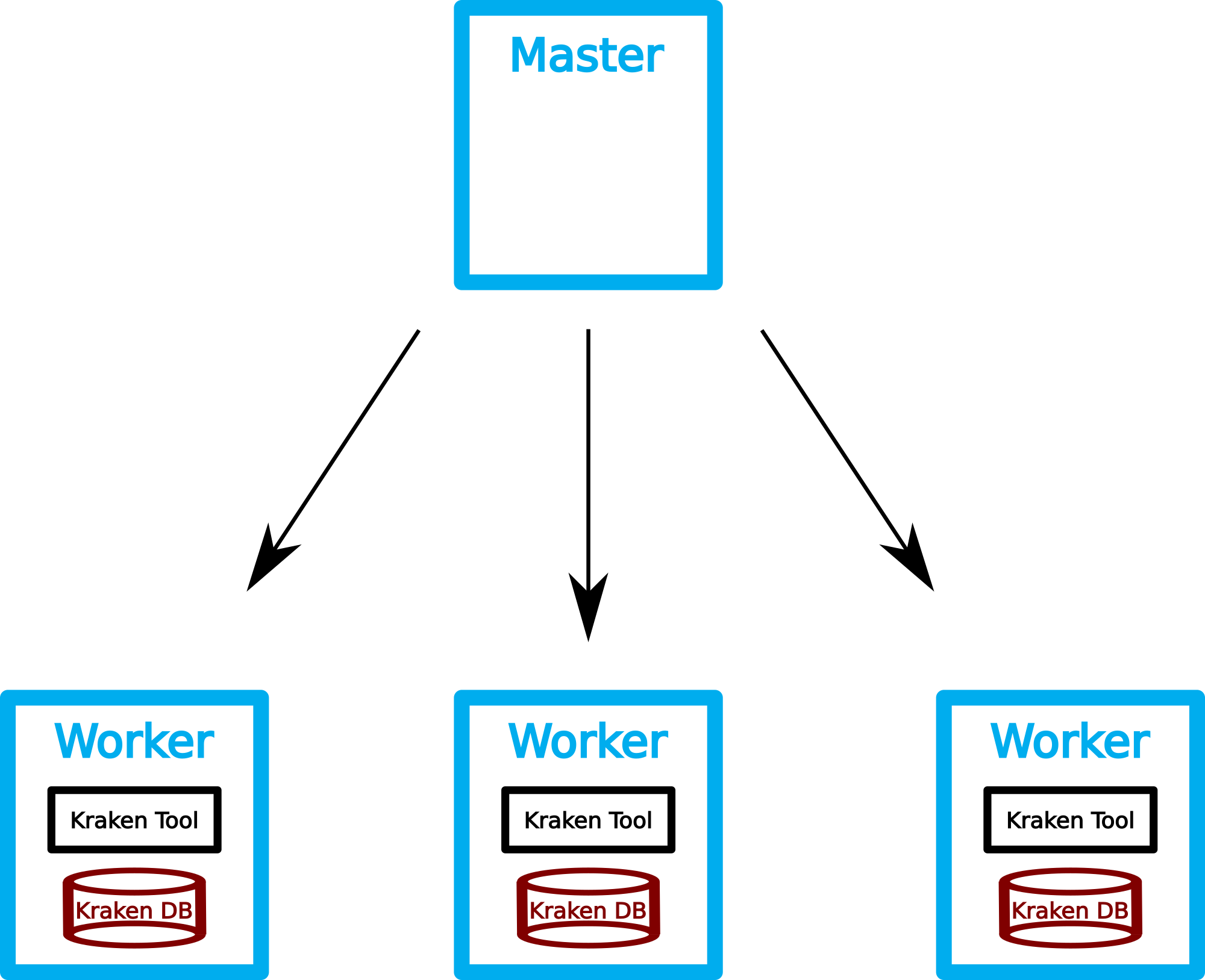
First we need to download the Kraken database to each of
the hosts. For usage with Kraken the database must decompressed before usage. Both actions are implemented in a shell script named kraken_download_db.sh.
We again use the GridEngine to distribute the script on all slave hosts.
cd ~/kraken-demo
qsub -t 1-4 -pe multislot 16 kraken_download_db.shFor the kraken analysis we have to use a shell script kraken_pipline.sh that should do the following :
-
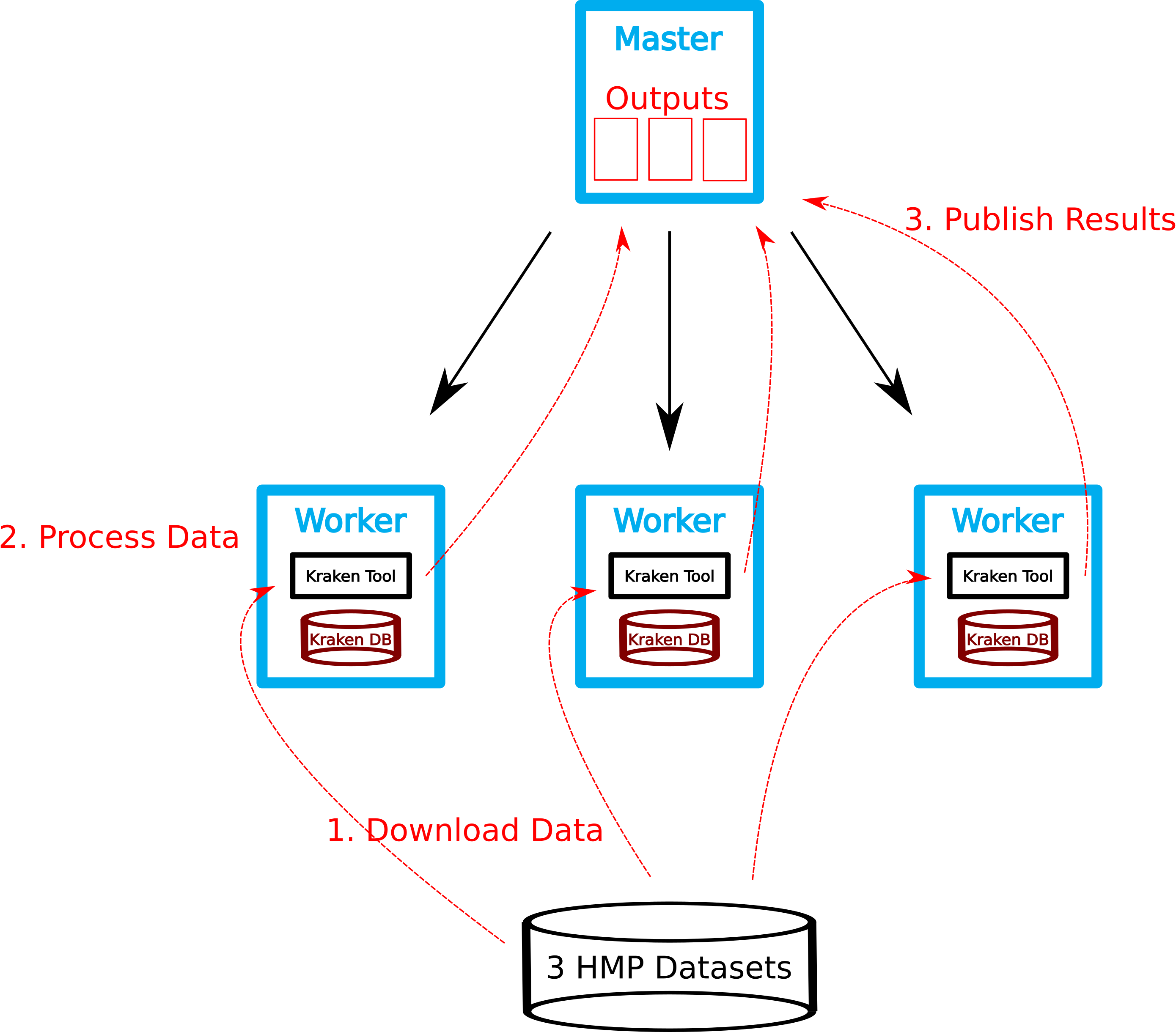
Download FASTQ file.
- the object storage contains a set of fastq files.
- Use
wgetto download them to local storage (/vol/scratch)
-
Run Kraken.
- FastQ files are compressed (tar.bz2) :
--fastq-input - mount input data and database path into the container
- Create Kraken report.
cd ~/kraken-demo
SEQ_FILES=$(curl -s https://openstack.cebitec.uni-bielefeld.de:8080/swift/v1/eMed/ | grep tar.bz2 )
for SF in ${SEQ_FILES}; do
SRS_NR=$(echo $SF | cut -f 2 -d '/' | cut -f 1 -d '.' );
qsub -pe multislot 16 kraken_pipeline.sh $SF $SRS_NR.report ;
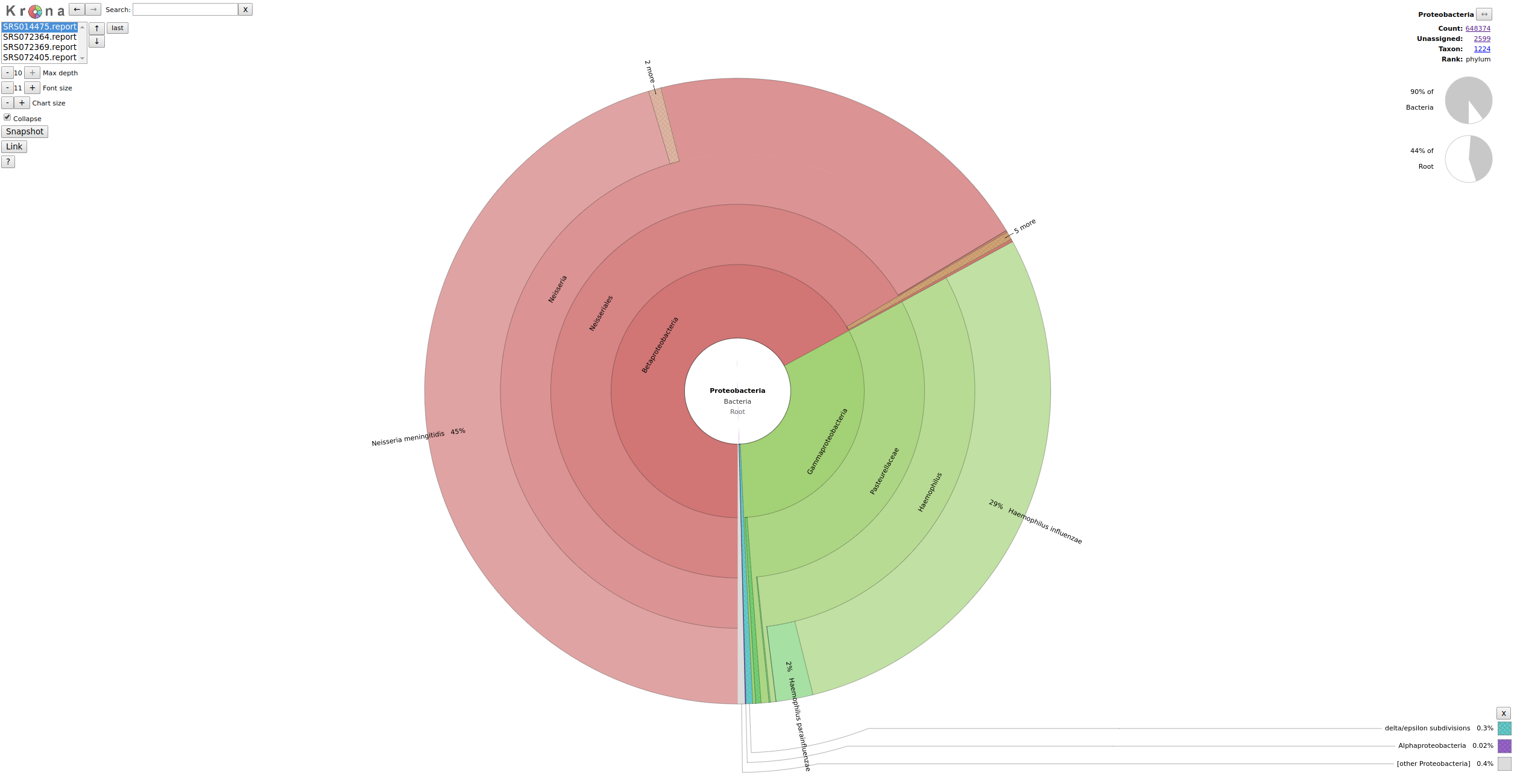
done We now use Krona to get a nice visualitation of our results. Again search for a suitable container containing the Krona software suite or build a Krona container by yourself, the hands-on repository contains a suitable Dockerfile.
Attention: Unfortuneatly the krona container offered by biocontainers.pro does not come with a preinstalled taxonomy database and the contained updateTaxonomy.sh script breaks because of a missing dependency (curl).
cd ~/kraken-demo
docker build -t krona .
Since all reports files located on a shared filesystem, Krona can be run directly on the master. We have to merge all kraken-reports ...
cd /vol/spool/kraken
for i in *.report.out; do cut -f2,3 $i > $i.krona; done
cd /vol/spool/kraken
docker run -u 1000:1000 -v /vol/spool/kraken:/data krona ktImportTaxonomy *.krona -o krona.html
sudo mv /vol/spool/kraken/krona.html* /var/www/html
You can use your browser to look at the Krona output.
http://<BIBIGRID_MASTER_IP>/krona.html