Code for MLHC 2021 Multi-Label Generalized Zero Shot Learning for the Classification of Disease in Chest Radiographs
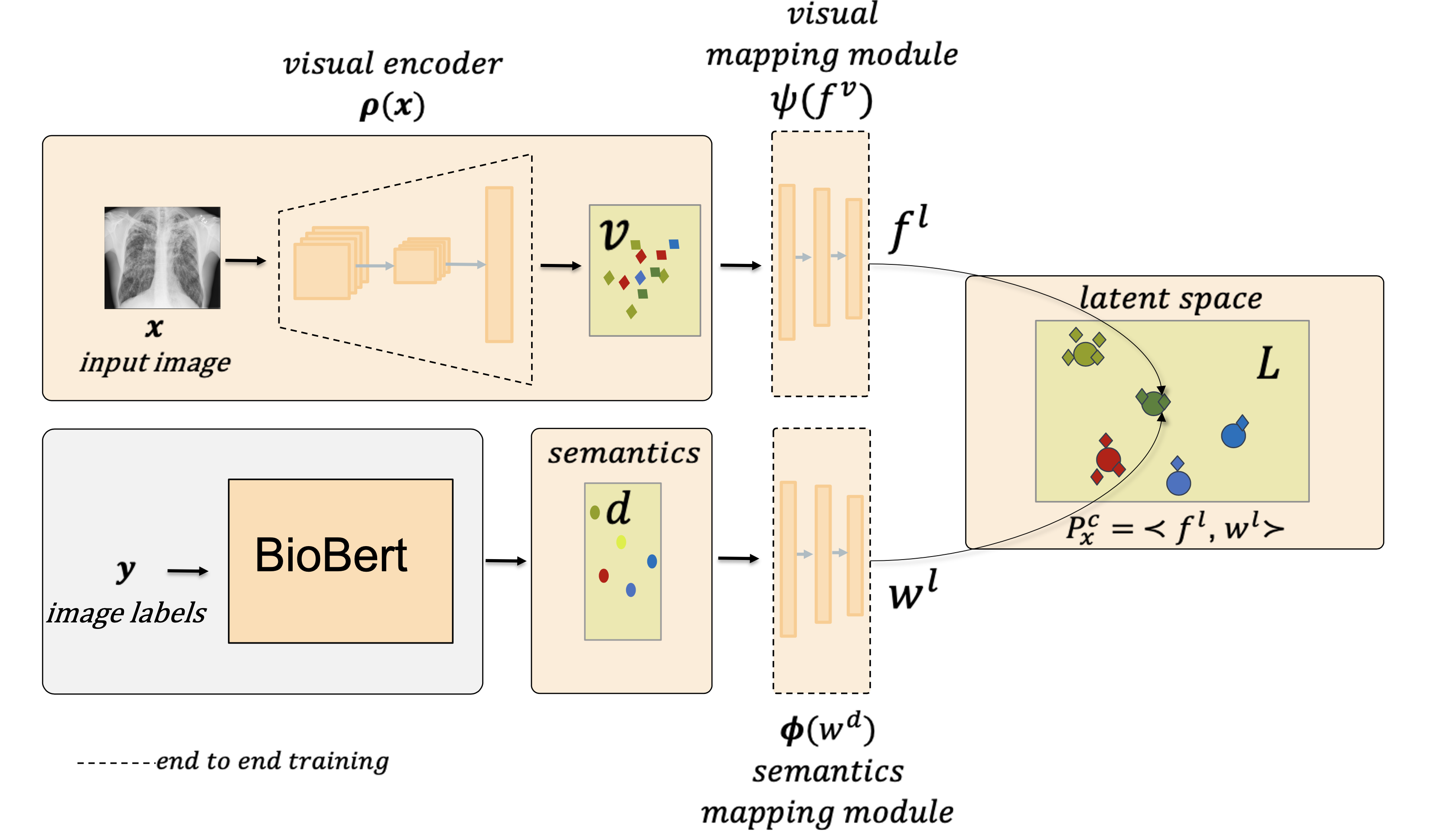
Despite the success of deep neural networks in chest X-ray (CXR) diagnosis, supervised learning only allows the prediction of disease classes that were seen during training. At inference, these networks cannot predict an unseen disease class. Incorporating a new class requires the collection of labeled data, which is not a trivial task, especially for less frequently-occurring diseases. As a result, it becomes inconceivable to build a model that can diagnose all possible disease classes. This repo contains the pytorch implementation for our proposed network; multi-label generalized zero shot learning (CXR-ML-GZSL) that can simultaneously predict multiple seen and unseen diseases in CXR images. Given an input image, CXR-ML-GZSL learns the visual representations guided by the input's corresponding semantics extracted from a rich medical text corpus.
The components of the network consist of (i) a trainable visual encoder, (ii) a fixed semantic encoder, & (iii) a projection module to map the encoded features to a joint latent space. Our approach is end-to-end trainable and does not require offline training of the visual feature encoder.
git clone https://github.com/nyuad-cai/CXR-ML-GZSL.git
cd CXR-ML-GZSL
conda env create -f environment.yml
conda activate zsl
We evaluated the proposed method on the NIH Chest X-ray dataset with a random split of 10 seen and 4 unseen classes. To train and evaluate the network, download the NIH chest x-ray dataset.
-
To perform evaluation only, you can simply download the pretrained network weights.
-
Update the paths of data-root directory and pretrained weights to run the following script.
sh ./scripts/test_densenet121.sh
-
To train the network, you run the following script by setting the data-root directory path of the downloaded dataset.
sh ./scripts/train_densenet121.sh
If you use this code for your research, please consider citing:
@misc{hayat2021multilabel,
title={Multi-Label Generalized Zero Shot Learning for the Classification of Disease in Chest Radiographs},
author={Nasir Hayat and Hazem Lashen and Farah E. Shamout},
year={2021},
eprint={2107.06563},
archivePrefix={arXiv},
primaryClass={cs.CV}
}
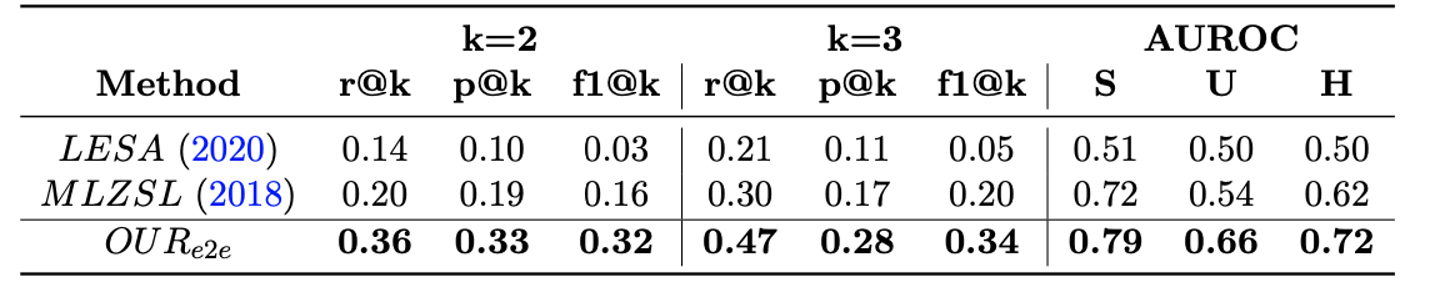
- We compare the performance of our proposed approach with two baseline methods and report overall precision, recall, f1-score @k {2,3} and AUROC for seen & unseen classes.
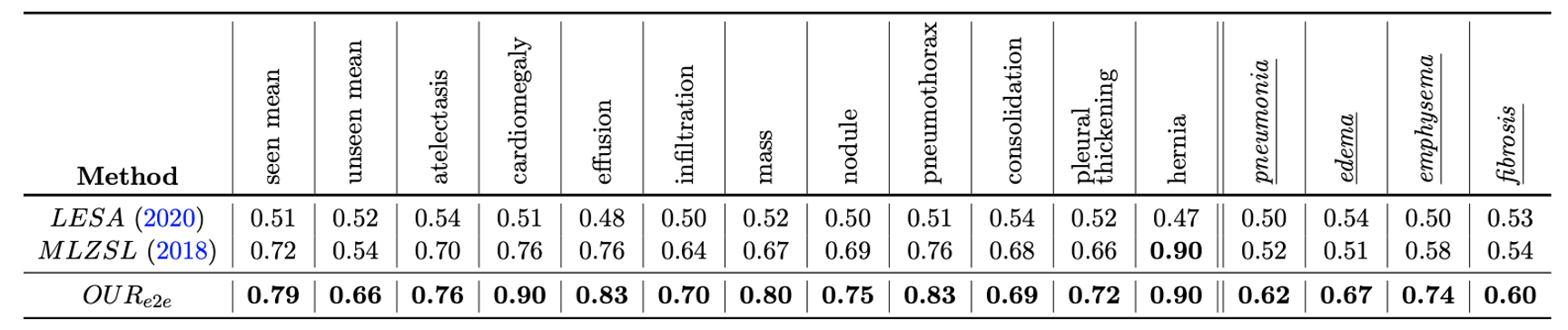
- Class wise comparison with baseline methods. The last 4 italicized classes are unseen during training.
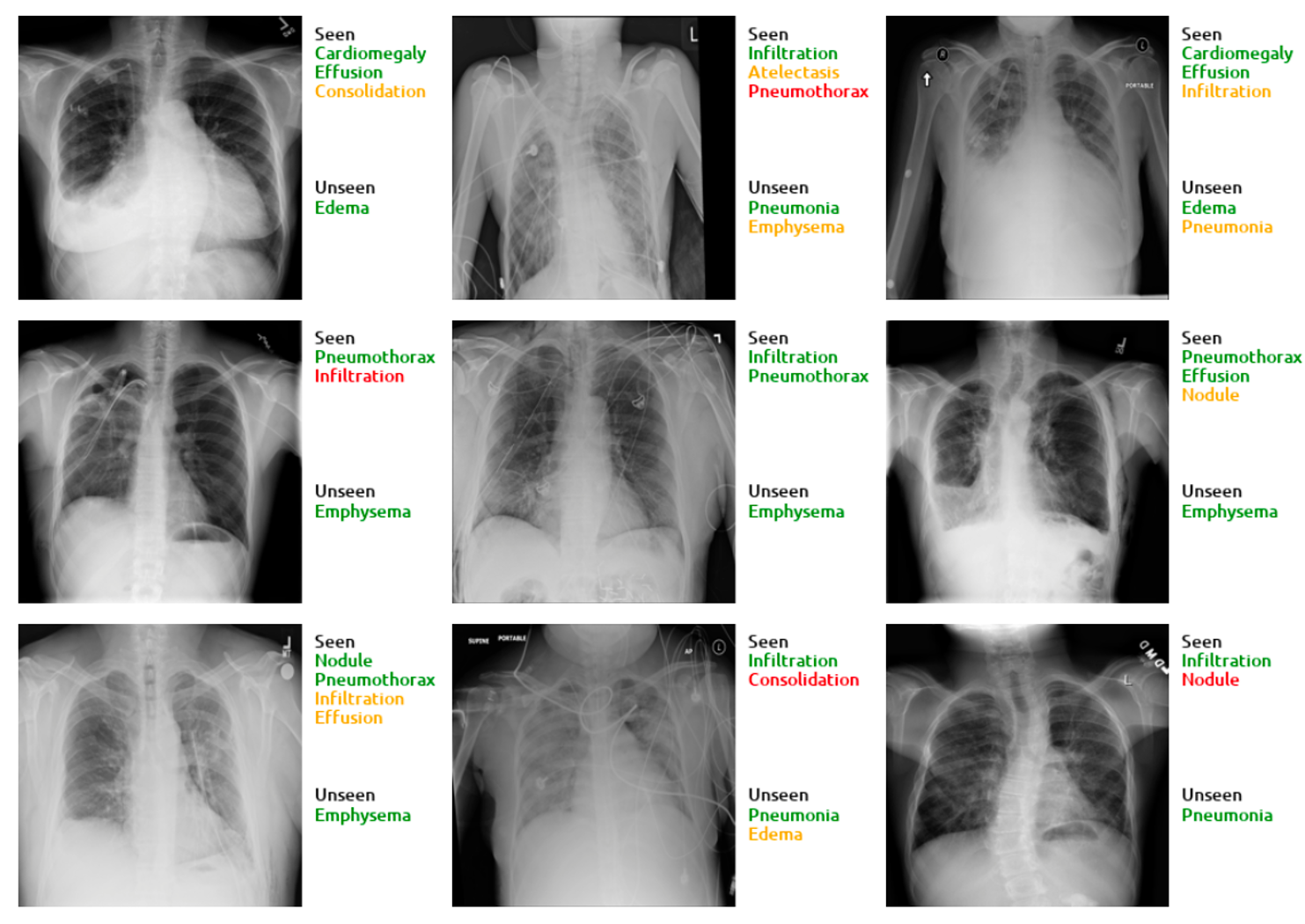
- Below we show some of the visual results for top 3 predictions. Green, orange, and red represent true positives, false negatives, and false positives, respectively. Note that our method predicts the unseen classes as top-3 even when the number of ground-truth labels is greater than 3.