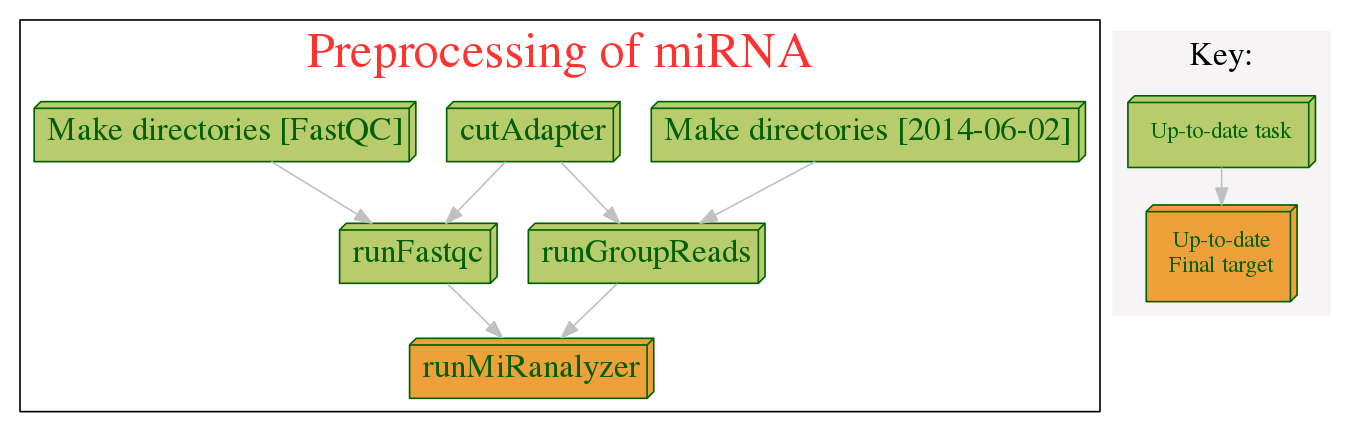
A pipeline of miRNA by using miRanalyzer
Here is the pipeline I use to analyze miRNA-seq data by miRanalyzer.
Now it support:
- cut adapter
- run FastQC as step of quality control
- align sequences to annotation databases
- predict novel miRNAs
- summarize the results and generate the count table of the entries
- The standalone version of miRanalyzer
- plyr An R package for data manipulation.
Install these softwares or packages and make sure the softwares are in $PATH.
Put all script in bin folders to a place in $PATH or add these folders to $PATH.
Firstly, you need to edit the config.yaml file to fit your need, then run:
nohup python pipline.py config.yaml &For the organization of projects, I generally follow this paper: A Quick Guide to Organizing Computational Biology Projects. So project_dir/data_dir/fastq are the folder contains raw fastq files, while project_dir/output_dir folder are the results. The position of scripts in project_script doesn't matter at all. But I prefer to put them under project/script/miRanalyzer folder.
When the major part of the pipeline finishs, then run:
Rscript mergeStats.R config.yaml
Rscript mergeTables.R config.yamlto summarize the final results. The items in stats_tags of config.yaml will be summarized.