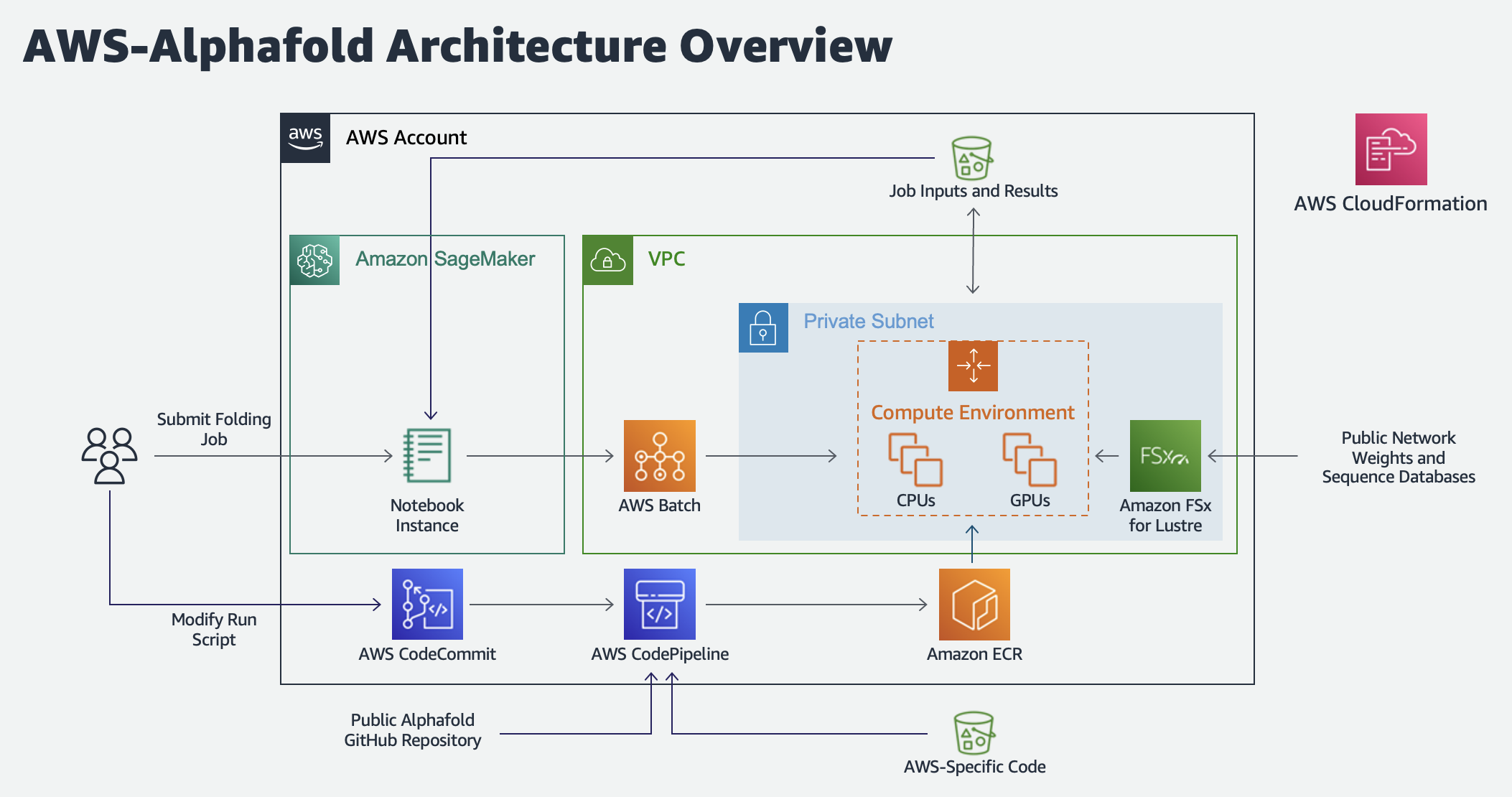
AWS Batch Architecture for AlphaFold
Overview
Proteins are large biomolecules that play an important role in the body. Knowing the physical structure of proteins is key to understanding their function. However, it can be difficult and expensive to determine the structure of many proteins experimentally. One alternative is to predict these structures using machine learning algorithms. Several high-profile research teams have released such algorithms, including AlphaFold 2, RoseTTAFold, and others. Their work was important enough for Science magazine to name it the "2021 Breakthrough of the Year".
Both AlphaFold 2 and RoseTTAFold use a multi-track transformer architecture trained on known protein templates to predict the structure of unknown peptide sequences. These predictions are heavily GPU-dependent and take anywhere from minutes to days to complete. The input features for these predictions include multiple sequence alignment (MSA) data. MSA algorithms are CPU-dependent and can themselves require several hours of processing time.
Running both the MSA and structure prediction steps in the same computing environment can be cost inefficient, because the expensive GPU resources required for the prediction sit unused while the MSA step runs. Instead, using a high-performance computing (HPC) service like AWS Batch allows us to run each step as a containerized job with the best fit of CPU, memory, and GPU resources.
This repository includes the CloudFormation template, Jupyter Notebook, and supporting code to run the Alphafold v2.0 algorithm on AWS Batch.
Architecture Diagram
First time setup
Deploy the infrastructure stack
- For Stack Name, enter a value unique to your account and region.
- For StackAvailabilityZone choose an availability zone.
- If needed, specify the following optional parameters:
- Select a different value for AlphaFoldVersion if you want to include another version of the public Alphafold repo in your Batch job container.
- Specify a different value for CodeRepoBucket and CodeRepoKey if you want to populate your AWS-Alphafold CodeCommit repository with custom code stored in another S3 bucket.
- Select a different value for FSXForLustreStorageCapacity if you want to provision a larger file system. The default value of 1200 GB is sufficient to store compressed instances of the Alphafold parameters, BFD (small and reduced), Mgnify, PDB70, PDB mmCIF, Uniclust30, Uniref90, UniProt, and PDB SeqRes datasets.
- Select a different value for for FSxForLustreThroughput if you have unique performance needs. The default is 500 MB/s/TB. Select a higher value for performance-sensitive workloads and a lower value for cost-sensitive workloads.
- Select Y for LaunchSageMakerNotebook if you want to launch a managed sagemaker notebook instance to quickly run the provided Jupyter notebook.
- Select I acknowledge that AWS CloudFormation might create IAM resources with custom names.
- Choose Create stack.
- Wait 30 minutes for AWS CloudFormation to create the infrastructure stack and AWS CodeBuild to build and publish the AWS-AlphaFold container to Amazon Elastic Container Registry (Amazon ECR).
Launch SageMaker Notebook
(If LaunchSageMakerNotebook set to Y)
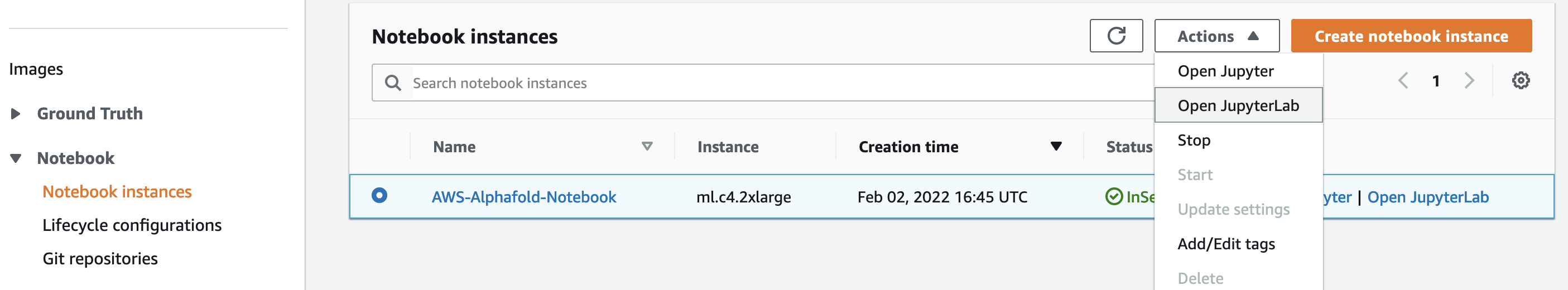
- Navigate to SageMaker
- Select Notebook > Notebook instances.
- Select the AWS-Alphafold-Notebook instance and then Actions > Open Jupyter or Open JupyterLab.
Clone Notebook Repository
(If LaunchSageMakerNotebook set to N)
- Navigate to CodeCommit.
- Select the aws-alphafold repository that was just created and copy the clone URL.
- Use the URL to clone the repository into your Jupyter notebook environment of choice, such as SageMaker Studio.
Populate FSx for Lustre File System
- Once the CloudFormation stack is in a CREATE_COMPLETE status, you can begin populating the FSx for Lustre file system with the necessary sequence databases. To do this automatically, open a terminal in your notebooks environment and run the following commands from the AWS-AlphaFold directory:
> pip install -r notebooks/notebook-requirements.txt
> python notebooks/download_ref_data.py --stack_name <STACK NAME>
Replacing <STACK NAME> with the name of your cloudformation stack. By default, this will download the "reduced_dbs" version of bfd. You can download the entire database instead by specifying the --download_mode full_dbs option.
- It will take several hours to populate the file system. You can track its progress by navigating to the file system in the FSx for Lustre console.
Cleaning Up
- To delete all provisioned resources from from your account, navigate to S3 and search for your stack name. This should return a bucket named
<STACK NAME>followed bycodepipelines3bucketa random key. Select the bucket and then Delete. - Navigate to ECR and search for your stack name. This should return two container repositories named
<STACK ID>followed by eitherdownloadcontainerregistryorfoldingcontainerregistry. Select both repositories and then Delete. - Finally, navigate to Cloud Formation, select your stack, and then Delete.
Usage
Use the provided AWS-AlphaFold.ipynb notebook to submit sequences for analysis and download the results.
Performance
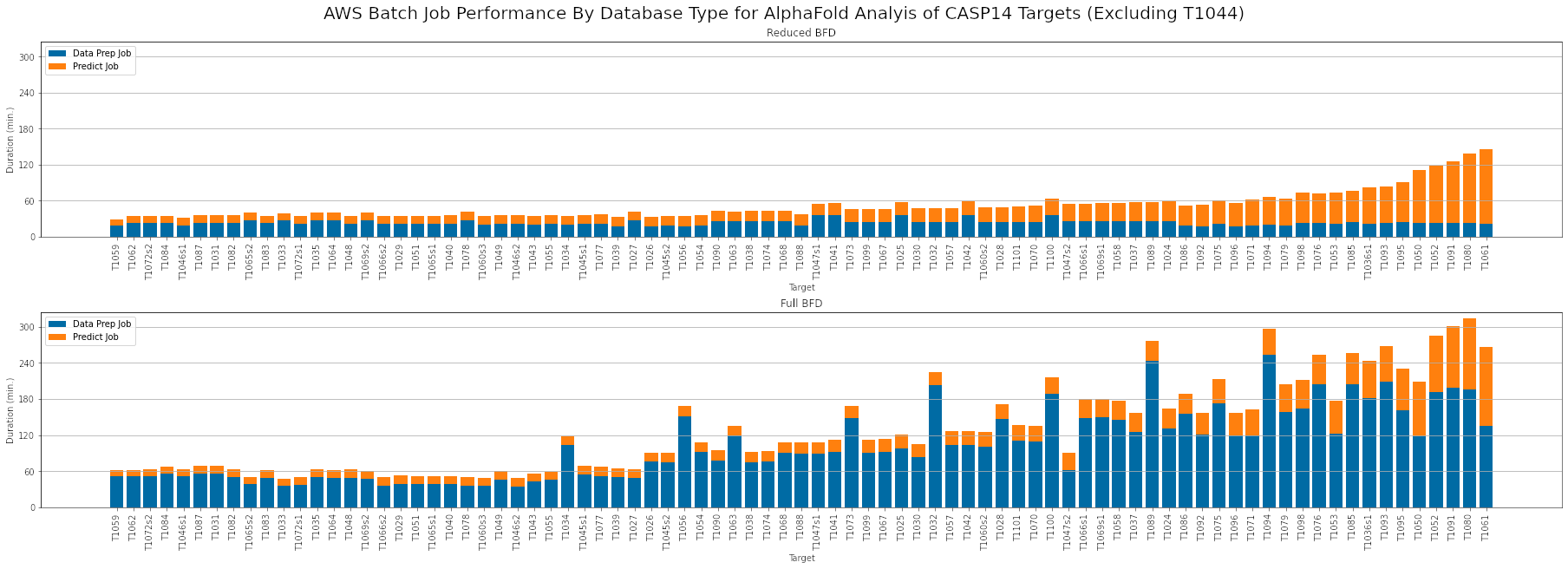
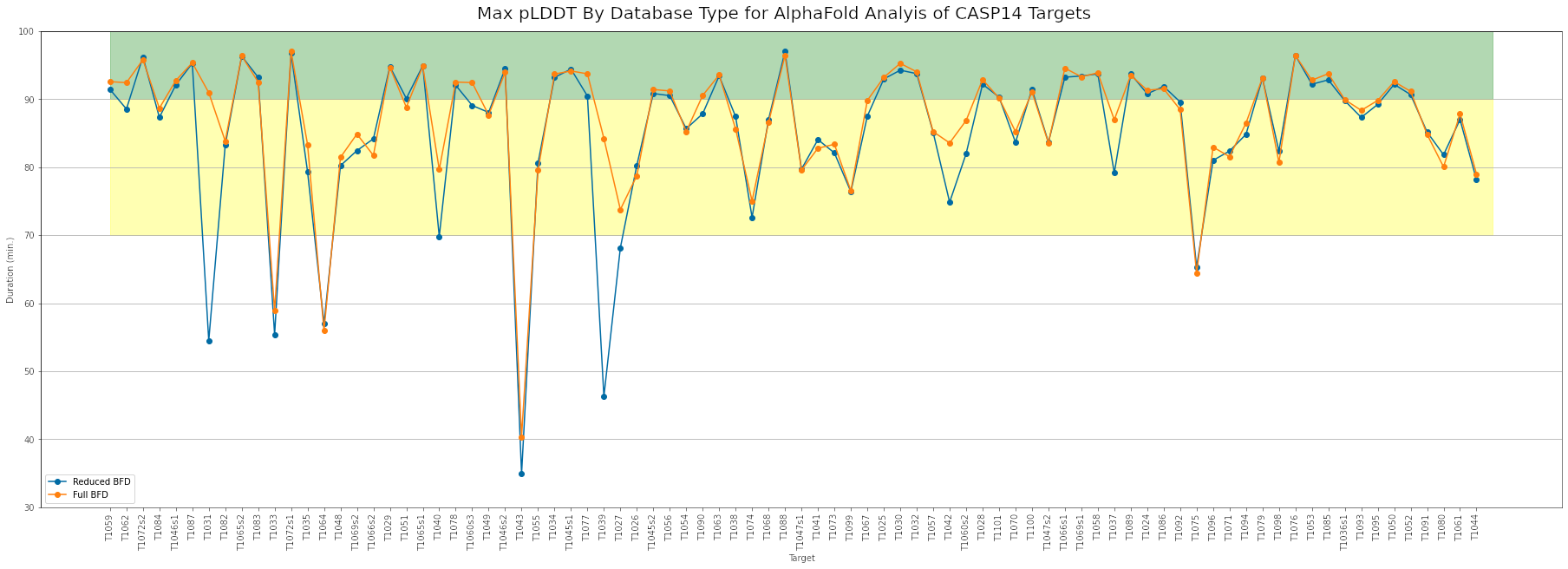
To determine the optimal compute settings, we used the CASP14 target list to test various CPU, memory, and GPU settings for the data preparation and prediction jobs. Using the full BFD database can increase the duration of data preparation jobs by as much as 10x. However the resulting increase in MSA coverage can increase the maximum pLDDT scores for some targets.
Based on this analysis, we recommend the following AWS Batch compute resource settings:
Data Preparation (Reduced BFD)
- CPsU: 4 vCPU
- Memory: 16 GiB
- GPUs: 0
Data Preparation (Full BFD)
- CPUs: 16 vCPU
- Memory: 32 GiB
- GPUs: 0
Prediction (Sequence Length < 700)
- CPUs: 4
- Memory: 16 GiB
- GPUs: 1
Prediction (Sequence Length > 700)
- CPUs: 16
- Memory: 64 GiB
- GPUs: 1
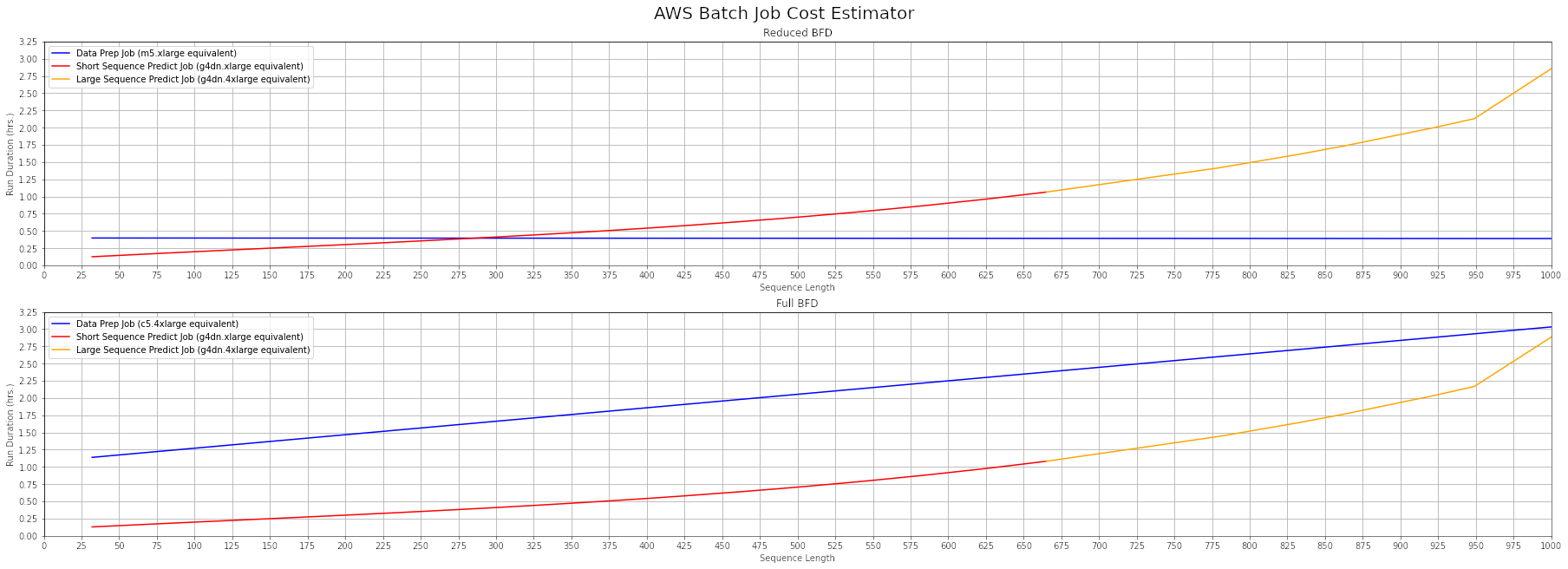
Cost Estimation
Follow these steps to estimate the per-run costs asociated with a protein of size X:
- Pick either the full or reduced bfd database
- Find the length of your target sequence on the x-axis.
- Use the plotted curves to identify the estimated data prep and prediction job durations.
- Refer to the EC2 On-Demand pricing page to obtain the hourly rate for the data prep job instance type equivalent (m5.xlarge or c5.4xlarge, depending on bfd database type) and prediction job instance type equivalent (g4dn.xlarge or g4dn.4xlarge, depending on sequence length).
For example, analyzing a 625-residue sequence using the reduced bfd database will take approximately 0.3 hours of data prep time, plus 1 hour of prediction time. As of February 2022, the on-demand rate for a m5.xlarge instance in the us-east-1 Region is $0.192/hr. and the rate for a g4dn.xlarge instance is $0.526/hr., for a total estimated cost of $0.72 per run. Please note that this pricing is subject to change at any time.
Security
See CONTRIBUTING for more information.
License
This project is licensed under the Apache-2.0 License.
Additional Information
AlphaFold Repository
Please visit https://github.com/deepmind/alphafold for more information about the AlphaFold algorithm.
Citations
The original AlphaFold 2 paper is
@Article{AlphaFold2021,
author = {Jumper, John and Evans, Richard and Pritzel, Alexander and Green, Tim and Figurnov, Michael and Ronneberger, Olaf and Tunyasuvunakool, Kathryn and Bates, Russ and {\v{Z}}{\'\i}dek, Augustin and Potapenko, Anna and Bridgland, Alex and Meyer, Clemens and Kohl, Simon A A and Ballard, Andrew J and Cowie, Andrew and Romera-Paredes, Bernardino and Nikolov, Stanislav and Jain, Rishub and Adler, Jonas and Back, Trevor and Petersen, Stig and Reiman, David and Clancy, Ellen and Zielinski, Michal and Steinegger, Martin and Pacholska, Michalina and Berghammer, Tamas and Bodenstein, Sebastian and Silver, David and Vinyals, Oriol and Senior, Andrew W and Kavukcuoglu, Koray and Kohli, Pushmeet and Hassabis, Demis},
journal = {Nature},
title = {Highly accurate protein structure prediction with {AlphaFold}},
year = {2021},
volume = {596},
number = {7873},
pages = {583--589},
doi = {10.1038/s41586-021-03819-2}
}
The AlphaFold-Multimer paper is
@article {AlphaFold-Multimer2021,
author = {Evans, Richard and O{\textquoteright}Neill, Michael and Pritzel, Alexander and Antropova, Natasha and Senior, Andrew and Green, Tim and {\v{Z}}{\'\i}dek, Augustin and Bates, Russ and Blackwell, Sam and Yim, Jason and Ronneberger, Olaf and Bodenstein, Sebastian and Zielinski, Michal and Bridgland, Alex and Potapenko, Anna and Cowie, Andrew and Tunyasuvunakool, Kathryn and Jain, Rishub and Clancy, Ellen and Kohli, Pushmeet and Jumper, John and Hassabis, Demis},
journal = {bioRxiv}
title = {Protein complex prediction with AlphaFold-Multimer},
year = {2021},
elocation-id = {2021.10.04.463034},
doi = {10.1101/2021.10.04.463034},
URL = {https://www.biorxiv.org/content/early/2021/10/04/2021.10.04.463034},
eprint = {https://www.biorxiv.org/content/early/2021/10/04/2021.10.04.463034.full.pdf},
}