First, clone repository using git clone.
$ git clone https://github.com/noepinefrin/ABpy.gitThen, install the dependencies.
$ pip install -r requirements.txtNow, you can analysis what you want.
from sklearn.datasets import load_breast_cancer
import pandas as pd
brca = datasets.load_breast_cancer(as_frame=True)
brca_data = pd.concat([brca.data, brca.target], axis=1)from ab.ab import ABpy
brca_abpy = ABpy(data=brca_data, target_class='target', test_variables=brca.data.columns, significance_level=0.05)
brca_ab_results = brca_abpy.apply(verbose=False) # returns pd.DataFrame, when you are using verbose=True, you also get the interpretation of the variables.brca_ab_results.head(3)| Distribution of A | Distribution of B | Shapiro P-Value of A | Shapiro P-Value of B | Shapiro W-Value of A | Shapiro W-Value of B | Equal Variance | Levene P-Value | Levene F-Value | T-Test P-Value | T-Test T-Value | Mann Whitney U-Test P-Value | Mann Whitney U-Test U-Value | Mean Ratio | Median Ratio | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Feature 1 | normal | normal | 5.554e-01 | 2.610e-01 | 0.980081 | 0.97129 | equal-variance | 8.585e-02 | 3.010904 | 3.126e-10 | -7.005796 | - | - | 1.513633 | - |
| Feature 2 | non-normal | normal | 7.264e-03 | 5.375e-01 | 0.933116 | 0.979655 | - | - | - | - | - | 1.743e-04 | 1795.0 | - | 1.245406 |
| Feature 3 | non-normal | non-normal | 2.309e-02 | 5.364e-03 | 0.945846 | 0.983864 | - | - | - | - | - | 2.142e-06 | 1938.0 | - | 3.283711 |
brca_ab_results = brca_abpy.apply(verbose=True)+ A/B Testing for FEATURE1
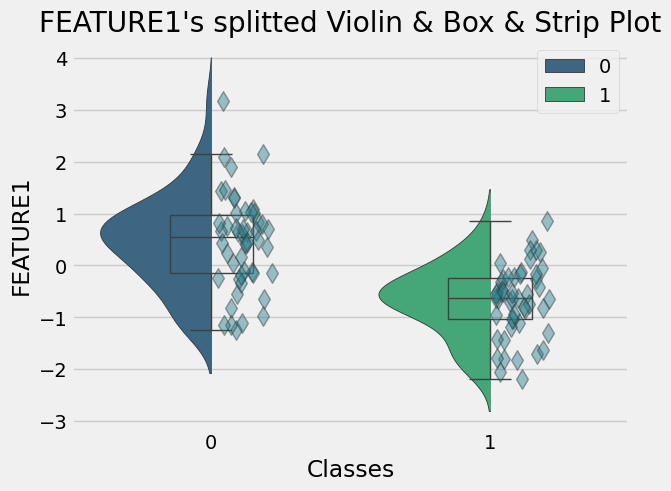
- Summary Statistics by Groups for FEATURE1
target 0 1
count 50.000000 50.000000
mean 0.447790 -0.677789
std 0.913841 0.674936
median 0.541742 -0.621856
min -1.235532 -2.181159
max 3.167797 0.849184
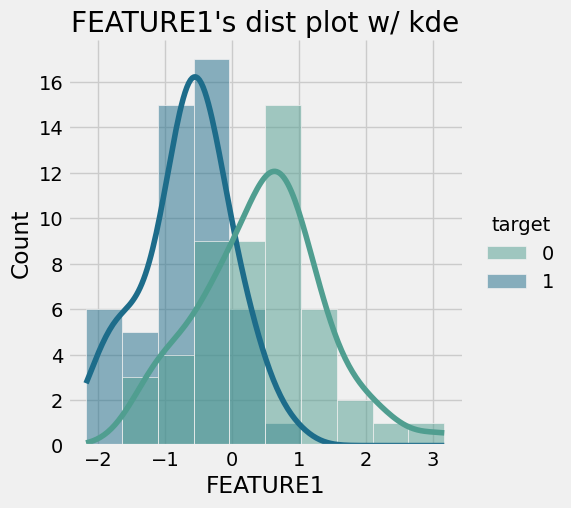
- Histogram by groups for FEATURE1- Violin-Box-Strip Plot by groups for FEATURE1+ 1. Step: Testing the Normality Assumption for FEATURE1 using Shaphiro Wilk Test
A P-Value: 5.554e-01
B P-Value: 2.610e-01
Shaphiro Wilk Test resulted as p > 0.05 for A and B which indicates that H0 can NOT be rejected.
Accordingly distribution of FEATURE1 values in A and B are likely to normal distribution.
+ 2. Step: Testing the Homogeneity Assumption for FEATURE1 using Levene's F-Test
Levene P-Value: 8.585e-02 & B Levene F-Value: 3.0109044461371997
Levene's F-Test resulted as p > 0.05 for A and B which indicates that H0 can NOT be rejected.
Accordingly variance of FEATURE1 values in A and B are equal.
+ 3. Step: Independent samples T-Test for FEATURE1 using T-Test
T-Test P-Value: 3.126e-10 & T-Test T-Value: -7.005795820145608
Independent samples T-Test resulted as p < 0.05 for A and B which indicates that H0 is rejected.
Accordingly T-Test results, there is significant difference between A and B for FEATURE1.
Mean of B in FEATURE1 is greater than A.For any inquiries or questions regarding ABpy or its functionalities, please feel free to contact me directly. I can be reached via email at berkayozcelik77@hotmail.com or through my LinkedIn profile at LinkedIn.