(Selection by Iterative pathway Group and Network Analysis Looping)
The Selection by Iterartive pathway Group and Network Analysis Looping (SIGNAL) platform is designed to facilitate robust hit selection from high-throughput studies.
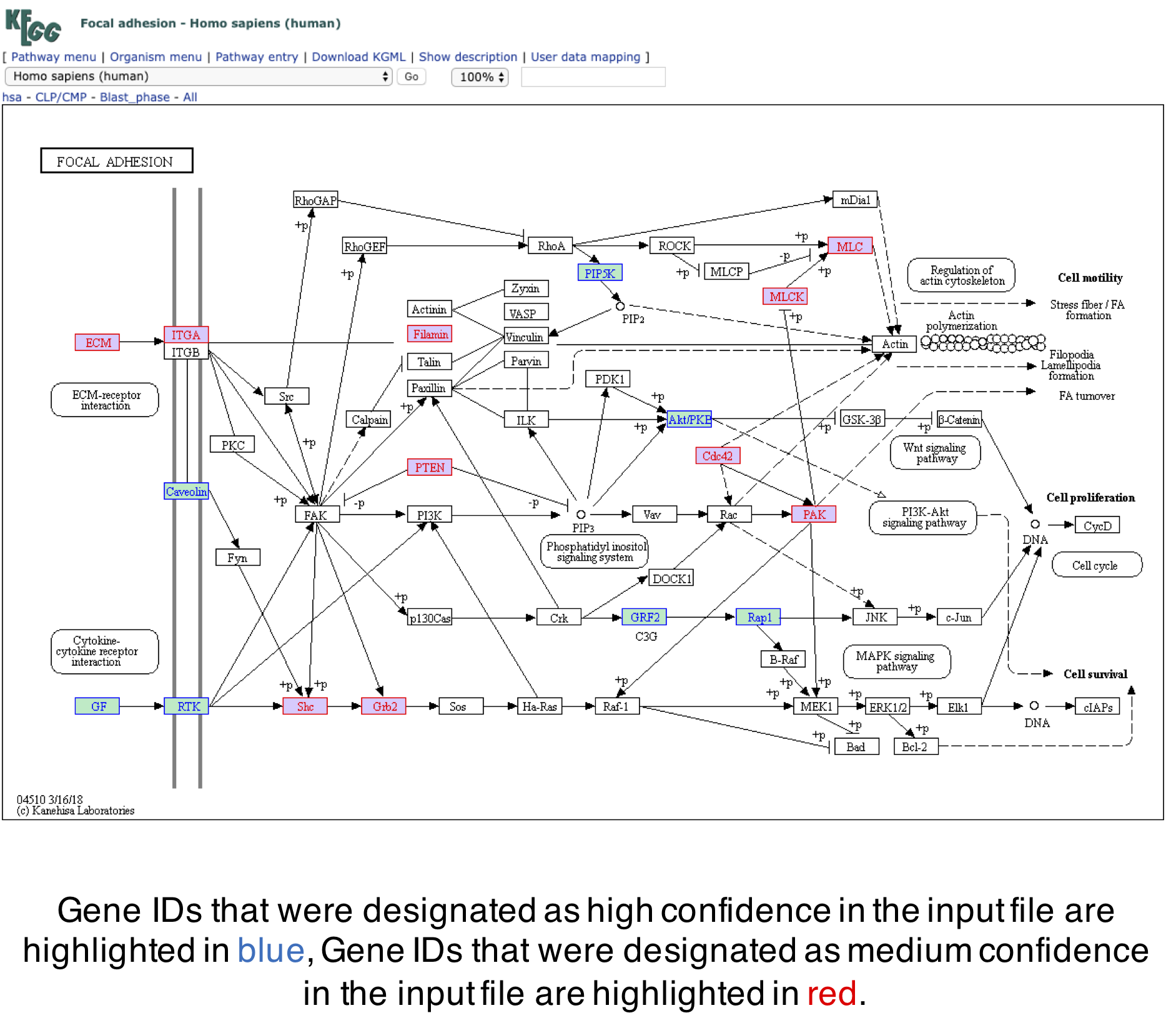
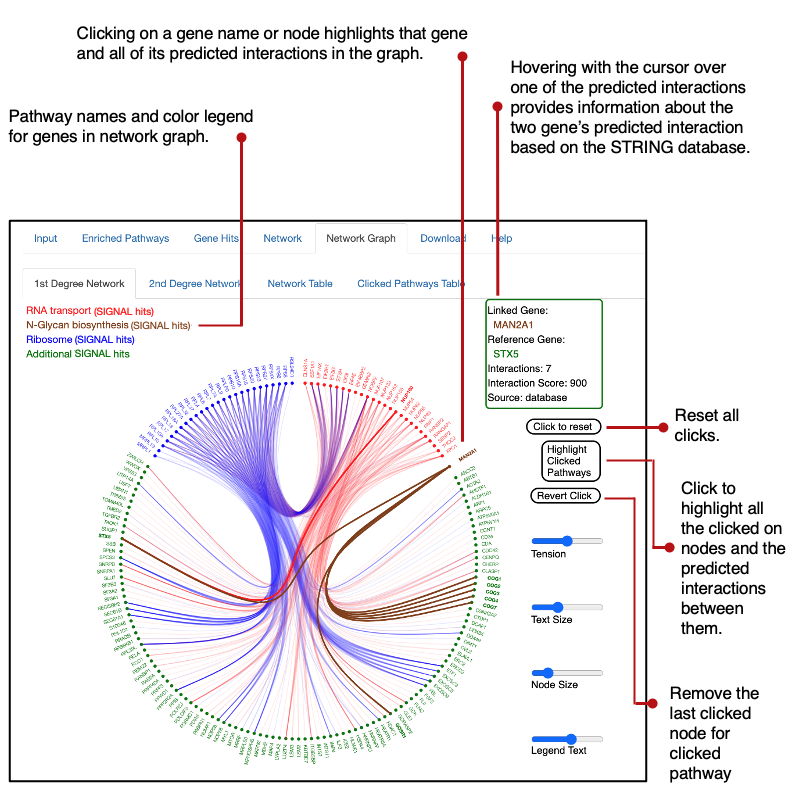
SIGNAL integrates analysis from protein to protein predicted interactions and statistical enrichments of known gene sets to correct for the false positive and false negative error rates that are associated with normalization methods that require arbitrary cutoffs. By utilizing data sorted into low, medium, and high confidence tiers the SIGNAL platform integrates the experimental readout of the analyzed study with curated knowledge of biological networks and interactions.
SIGNAL is best utilized as tool for selecting candidates from high-throughput studies to be further validated by rigorous low throughput follow up studies. The analysis outputs and platform features are designed to guide prediction, analysis, and hypothesis generation from large-scale datasets that can then be tested in targeted follow up assays.
SIGNAL is available as publicly facing website, as an R shiny app that can be run locally, and as a standalone R function.
SIGNAL is available on the Internet through our development site or/and production site. SIGNAL can also be downloaded and run as a standalone application. The instructions below will get you a copy of SIGNAL up and running on your local machine.
To run SIGNAL on your local machine, R and Rstudio should be installed and running on your machine. Instructions are available for R installation and for RStudio installation. In addition, several R packages are also required for SIGNAL to work on your local machine.
# To install required R packages from R commandline
library(shiny)
library(shinyjs)
library(shinyBS)
library(readr)
library(dplyr)
library(stringi)
library(DT)
library(data.table)
library(igraph)
library(edgebundleR)
library(shinyAce)
library(rJava)
library(networkD3)
library(visNetwork)
library(org.Hs.eg.db)
library(org.Mm.eg.db)
library(AnnotationDbi)
library(reshape2)
library(ggplot2)
library(tidyr)
library(gridExtra)
library(crosstalk)
library(htmltools)
library(stringr)
library(tools)
library(readxl)
Sys.setenv(R_ZIPCMD="/usr/bin/zip")
Before installing SIGNAL, you need to install git, if not installed already, by following these instructions. After installing git, you can install SIGNAL on your local machine in your home directory or anywhere under your home directory:
# For developmental version
$git clone https://github.niaid.nih.gov/Signaling-Systems-Unit/SIGNAL.git
# Or for production version
$git clone https://github.com/niaid/SIGNAL
After this, you should see a SIGNAL directory which contains all required files and data to run SIGNAL on your local machine.
To run SIGNAL on your local machine, start RStudio first, open 'app.R' file in RStudio from your SIGNAL directory, and then click 'Run App' button to start SIGNAL. You should see SIGNAL running in your default web browser.
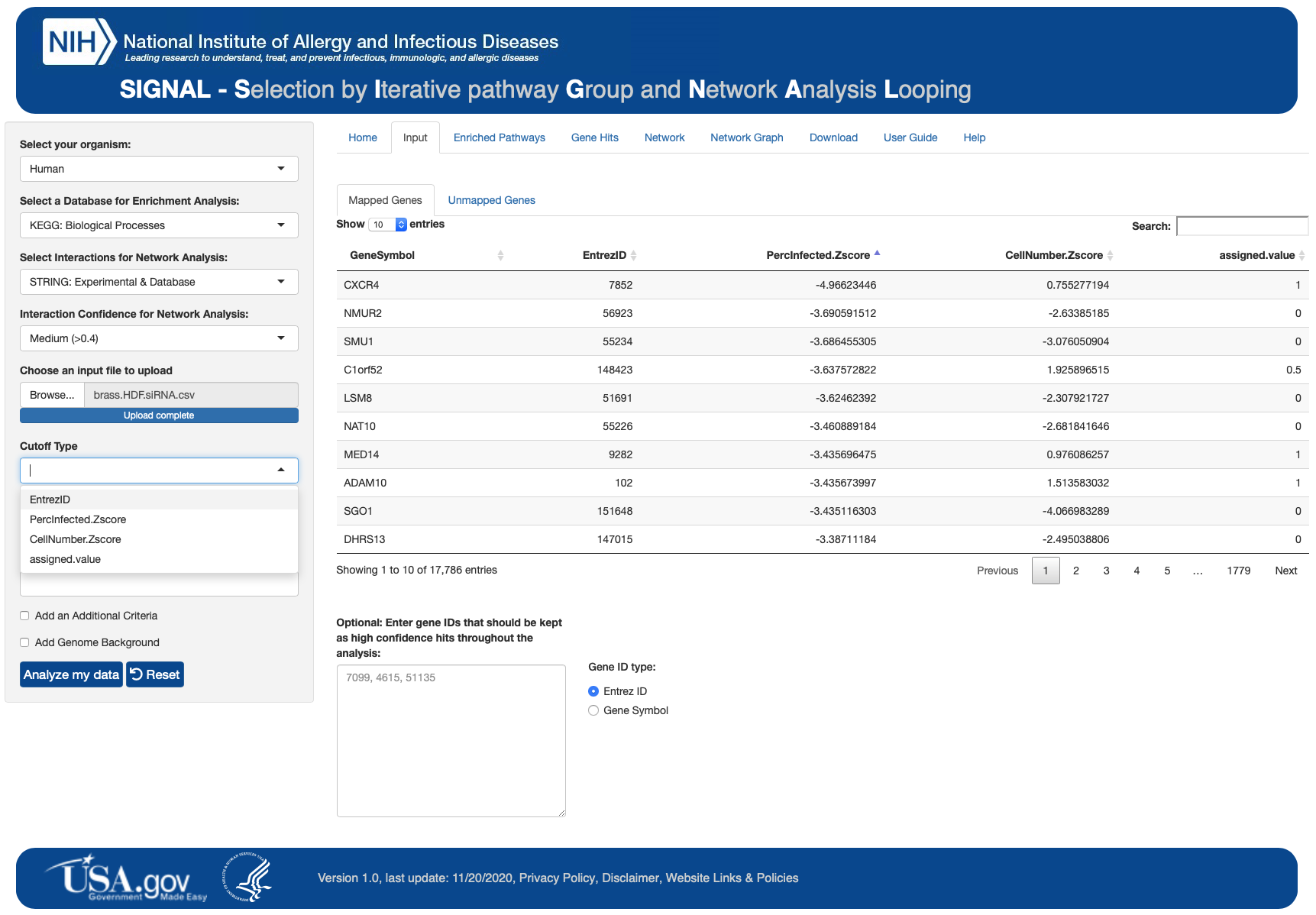
A sample input file (LPS_sample_dataset.csv) can be used to test SIGNAL. The sample file is a part of SIGNAL package and can be found in the SIGNAL directory:
# Sample input file
SIGNAL/app/www/SampleDatasets/LPS_sample_dataset.csv
A detailed user guide is available on website and in the SIGNAL directory:
# Detailed User Guide
SIGNAL/app/www/SIGNAL_userguide_V4.csv
To make SIGNAL an adaptable framework for iterative analysis with different datasets and databases beyond the databases and settings used on this platform, a R script version of a standalone SIGNAL function can be downloaded. The SIGNAL function relies on calling two separate analysis function, a pathway enrichment function and a network analysis function. The master SIGNAL function applies the pathway and network function iteratively, and the results are tested for when the analysis converges on a single set.
The list of input variables that can be selectively assigned in the adaptable SIGNAL function in R and their required formats are:
screen.datafame: A data frame of the screen.
ID.column: A column within the screen.dataframe for the identifiers of the targets (EntrezID, GeneSymbol, etc.).
criteria.column: A column within the screen.dataframe of the criteria for being considered a hit.
highconf.criteria: A criteria each target has to meet to be considered a "high confidence" hit.
midconf.criteria: A criteria each target has to meet to be considered a "mid confidence" hit.
criteria.setting: Whether the function should be using "equal", "greater than or equal", or "less than or equal" when assessing if confidence criteria is met. criteria.setting input should be in the format of "equal", "greater", or "less".
enrichment.dataframe: A data frame to be used for pathway membership in the format of a column of IDs (should be same as ID column in screen.dataframe in ID type and column title) and a column of which group they are part of. (each ID~group relationship needs to be in its own separate row).
enrichment.title: Name of the column with the names of the enrichment groups the targets are members of.
stat.test: Name of the statistical test to be used for measuring enrichment confidence. Needs to be in the format of either "pVal", "FDR", or "Bonferroni".
test.cutoff: A numeric value which a less than value in stat.test will be considered a significant enrichment. network.igraph: an igraph of the network to be used for network analysis (network igraph must use the same ID type as screen.dataframe)
The user provided variables are then used to apply the iterative function as in the previous paragraph. The adaptable version of SIGNAL broadens the possibility for its application beyond the use of the specific databases and settings it was designed in.
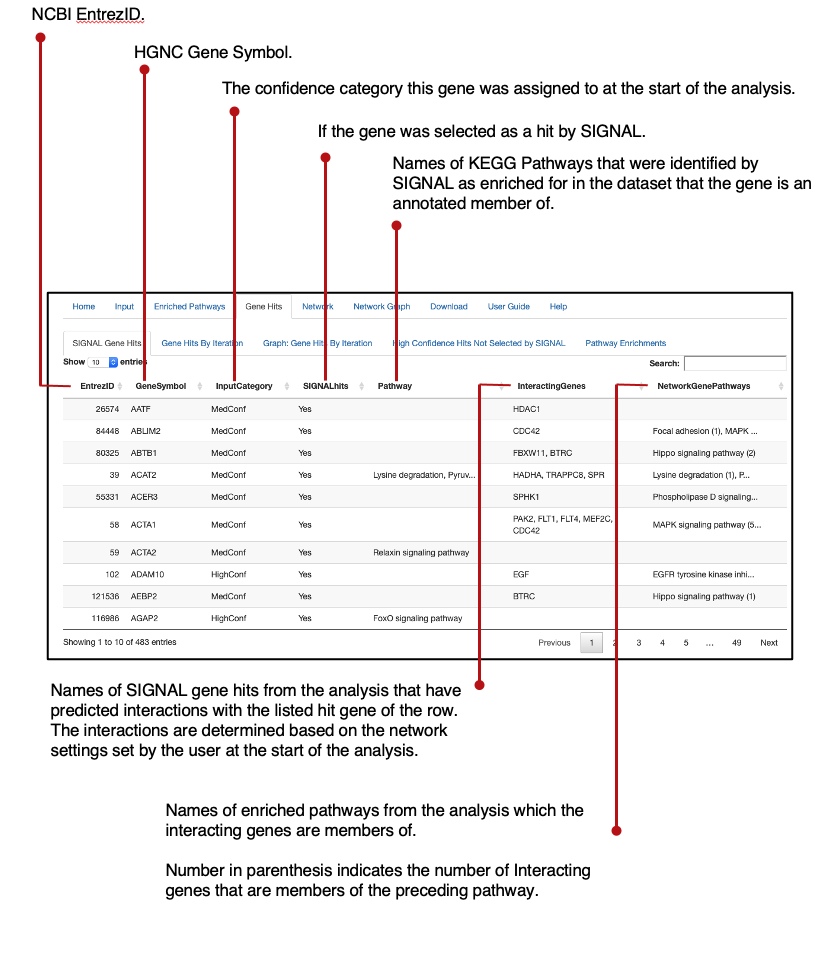
The SIGNAL function provides an output in the format of a R script list that contains three data frames:
- The input data frame with an appended 'SIGNAL.hit' column.
- A data frame of high confidence and medium confidence designation at each iteration of the analysis.
- A data frame of final SIGNAL enrichments from the provided enrichment data frame.
The SIGNAL function can be downloaded from the app
# SIGNAL as standalon R function
SIGNAL/app/www/RscriptsDownload/SIGNAL_R_function.R
The R code used to generate the analysis and figures for our manuscript is included in the github page.
# Manuscript R code
SIGNAL/manuscript/Rcode/
Sam Katz, Jian Song, Kyle Webb, Nicolas Lounsbury, Iain Fraser Contact us at signal-team@nih.gov
- We appreciate all the help we got from NIH\NIAID\OCICB, particularly the OEB Platform Team.
- We also want to thank our LSB colleagues at NIH/NIAID for their support.