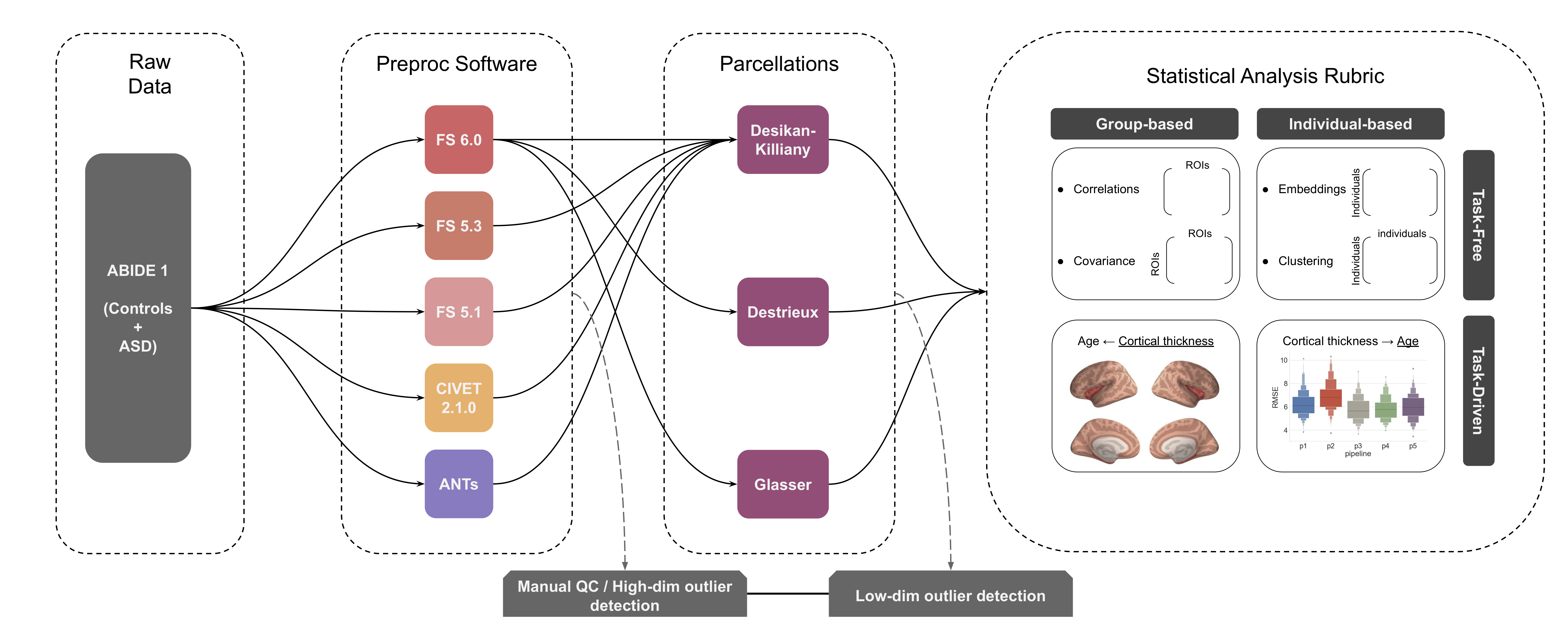
An extension of BrainHack Project (https://github.com/companat/compare-surf-tools) to compare thickness outputs from different pipelines run on ABIDE-I
- Compare output of preprocessing pipelines for structural MR imaging
- Software: Freesurfer (v5.1, v5.3, v6.0) ANTs, CIVET2.1
- Atlases: ROI-wise (surface parcellations: DKT40, Destrieux, Glasser)
- analytic comparisons: classifier performance (individual predictions), statistical inference (biological group differences)
- Quality control
- Manual / visual outlier detection
- Automatic outlier detection
Consolidated from the analysis results provided at http://preprocessed-connectomes-project.org/abide/Description. We provide the unified data tables in the 'data' directory. The subject lists for various analyses can be generated using software specific data tables and QC lists.
- ABIDE_Phenotype.csv : phenotypic data for the subjects
- ANTs, CIVET, FS* : preproc software output (ROI-wise)
- QC : QC lists from manual and automatic outlier detection
Legacy:
- see https://github.com/companat/compare-surf-tools for details
- notebooks: driver code to run analysis
- lib: helper functions for parsing and analysis
- scripts: code to read software output (civet2.1, fs6.0)
.
├── notebooks
│ ├── run_atlas_comparisons.ipynb
│ ├── run_software_comparisons.ipynb
│ ├── import_QC_data.ipynb
│ ├── Outlier_QC_analysis.ipynb
│ ├── generate_plots_individual_and_aggregates.ipynb
│ └── learn_pipeline_transforms.ipynb
└── lib
│ ├── data_handling.py
│ ├── deeplearning.py
│ ├── data_stats.py
│ └── plot_utils.py
└── scripts
├── get_vertex_data_fs.py
├── get_dkt_data_civet.py
├── get_roi_data_fs.py
└── check_vertex_data.py
Prereq: Processed output from a given software: e.g. FreeSurfer
A. Data parsing
- run scripts/get_vertex_data_fs.py on a FS subject dir to get vertext-wise summay CSV for all subjects.
python get_vertex_data_fs.py -s ../data/subjects/ -k '.fwhm20.fsaverage.mgh' -o ../data/sample_output/fs_fsaverage_vout
- run scripts/get_roi_data_fs.py on a FS subject dir to get ROI-wise summay CSV for all subjects. This script uses aparcstats2table command.
python get_roi_data_fs.py -s ../data/subjects -l ../data/subject_list.txt -m thickness -p a2009s -o ../data/sample_output/
B. Data standardization and comparative analyses
- run_atlas_comparisons.ipynb
- run_software_comparisons.ipynb
C. QC and outlier analysis
- import_QC_data.ipynb
- Outlier_QC_analysis.ipynb
D. Visualization of brainmaps
- generate_plots_individual_and_aggregates.ipynb