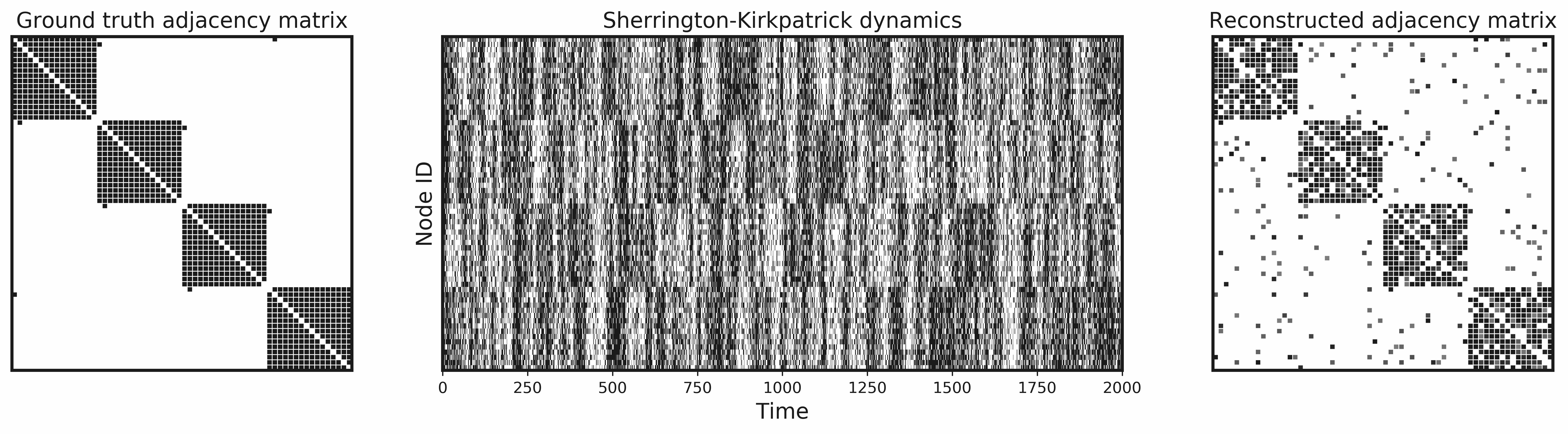
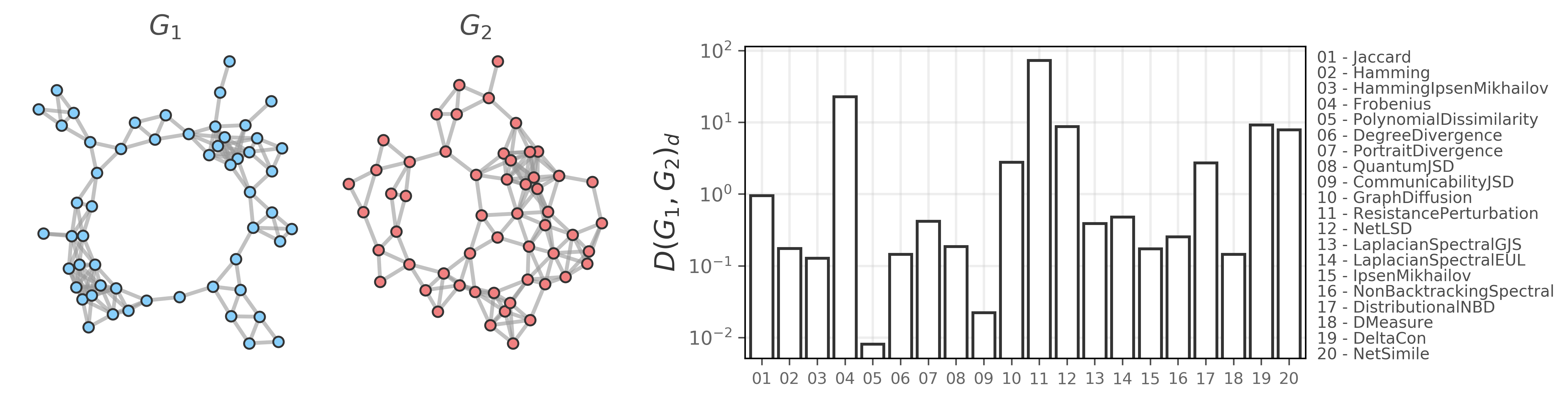
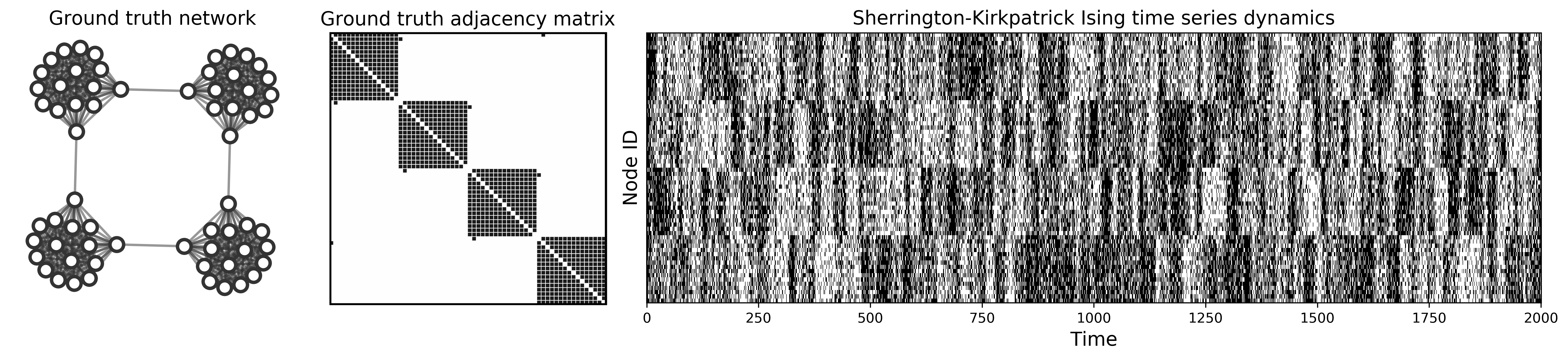
This library provides a consistent, NetworkX-based interface to various utilities for graph distances, graph reconstruction from time series data, and simulated dynamics on networks.
Some resources that maybe of interest:
netrd is easy to install through pip:
pip install netrd
If you are thinking about contributing to netrd, you can install a
development version by executing
git clone https://github.com/netsiphd/netrd
cd netrd
pip install .
The basic usage of a graph reconstruction algorithm is as follows:
from netrd.reconstruction import CorrelationMatrix
import numpy as np
# 100 nodes, 1000 observations
TS = np.random.random((100, 1000))
reconstructor = CorrelationMatrix()
G = reconstructor.fit(TS, threshold_type='degree', avg_k=15)
# or alternately, G = reconstructor.results['graph']Here, TS is an N x L numpy array consisting of L
observations for each of N sensors. This constrains the graphs
to have integer-valued nodes.
The results dict object, in addition to containing the graph
object, may also contain objects created as a side effect of
reconstructing the network, which may be useful for debugging or
considering goodness of fit. What is returned will vary between
reconstruction algorithms.
Many reconstruction algorithms create a dense matrix of weights and use additional parameters to describe how to create a sparse graph; the tutorial has more details on these parameters.
The basic usage of a distance algorithm is as follows:
from netrd.distance import QuantumJSD
import networkx as nx
G1 = nx.fast_gnp_random_graph(1000, .1)
G2 = nx.fast_gnp_random_graph(1000, .1)
dist_obj = QuantumJSD()
distance = dist_obj.dist(G1, G2)
# or alternatively: distance = dist_obj.results['dist']Here, G1 and G2 are nx.Graph objects (or subclasses such as
nx.DiGraph). The results dictionary holds the distance value, as
well as any other values that were computed as a side effect.
The basic usage of a dynamics algorithm is as follows:
from netrd.dynamics import VoterModel
import networkx as nx
ground_truth = nx.karate_club_graph()
dynamics_model = VoterModel()
synthetic_TS = dynamics_model.simulate(ground_truth, 1000)
# this is the same structure as the input data to a reconstructor
# G = CorrelationMatrix().fit(synthetic_TS)This produces a numpy array of time series data.
Contributing guidelines can be found in CONTRIBUTING.md.
-
McCabe, S., Torres, L., LaRock, T., Haque, S. A., Yang, C.-H., Hartle, H., and Klein, B. (2021). netrd: A library for network reconstruction and graph distances. Journal of Open Source Software 6(62): 2990. doi: 10.21105/joss.02990. arXiv: 2010.16019.
- paper detailing the methods used in this package
-
Hartle H., Klein B., McCabe S., Daniels A., St-Onge G., Murphy C., and Hébert-Dufresne L. (2020). Network comparison and the within-ensemble graph distance. Proceedings of the Royal Society A 476: 20190744. doi: 10.1098/rspa.2019.0744. arXiv: 2008.02415.
- recent work introducing a baseline measure for comparing graph distances