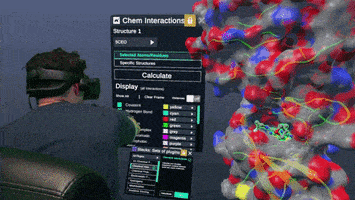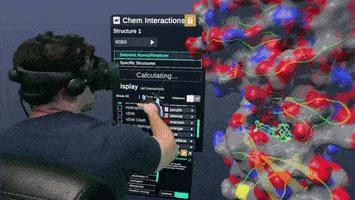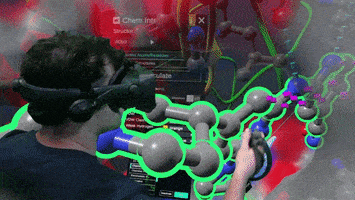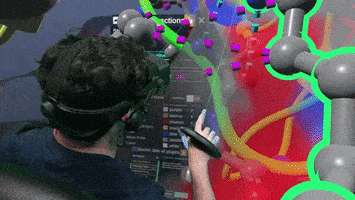
A Nanome plugin to Calculate and visualize interatomic contacts between small and macro molecules.
- Docker (https://docs.docker.com/get-docker/)
- (Optional) Docker Compose (https://docs.docker.com/compose/install/)
To start the plugin with production settings, we have a script available at ./docker/deploy.sh
./docker/build.sh
./docker/deploy.sh <args>There's two methods of configuring your plugin.
All Nanome plugins can be configured using the following set of command line args.
user@localhost:~/plugin-chemical-interactions$ python run.py --help
usage: run.py [-h] [-a HOST] [-p PORT] [-r] [-v] [-n NAME [NAME ...]]
[-k KEYFILE] [-i IGNORE]
Parse Arguments to set up Nanome Plugin
optional arguments:
-h, --help show this help message and exit
-a HOST, --host HOST connects to NTS at the specified url or IP address
-p PORT, --port PORT connects to NTS at the specified port
-r, --auto-reload Restart plugin automatically if a .py or .json file in
current directory changes
-v, --verbose enable verbose mode, to display Logs.debug
-n NAME [NAME ...], --name NAME [NAME ...]
Name to display for this plugin in Nanome
-k KEYFILE, --keyfile KEYFILE
Specifies a key file or key string to use to connect
to NTS
-i IGNORE, --ignore IGNORE
To use with auto-reload. All paths matching this
pattern will be ignored, use commas to specify
several. Supports */?/[seq]/[!seq]All of these args can be passed to docker/deploy.sh, and will be passed to the plugin container to connect your plugin to the correct NTS.
For example
./docker/deploy.sh -a nts-foobar.example.com -p 5555
When ./docker/deploy.sh is run, the command is copied into docker/redeploy.sh which can be used to redeploy your application without remembering the provided flags.
Alternatively, the chemical-interactions plugin supports storing NTS credentials in a .env file.
First thing you need to do is create a .env file in the top-level directory, which contains NTS connection information
NTS_HOST=foobar.example.com
NTS_PORT=5555
And then running the plugin is as simple as
./docker/deploy.sh --env-file <path to .env file> <plugin_args>Note that env files can be used alongside plugin args, but -a and -p will always take precedence over NTS_HOST and NTS_PORT
The plugin folder contains the entirety of the application.
- Handles all interactions with Nanome application
- Renders menus
- Visualizes interactions data in VR.
There's a separate conda environment installed in the container, where arpeggio is executed.
- Returns JSON containing interaction results, which is parsed by the plugin.
- See https://github.com/PDBeurope/arpeggio for more.
docker-compose.yml is optimized for development, with debug enabled and the code mounted as volumes.
If you use the VSCode IDE, we provide a .devcontainer, and debug launch configurations to ease development.
MIT
Harry C Jubb, Alicia P Higueruelo, Bernardo Ochoa-Montaño, Will R Pitt, David B Ascher, Tom L Blundell, Arpeggio: A Web Server for Calculating and Visualising Interatomic Interactions in Protein Structures. Journal of Molecular Biology, Volume 429, Issue 3, 2017, Pages 365-371, ISSN 0022-2836,