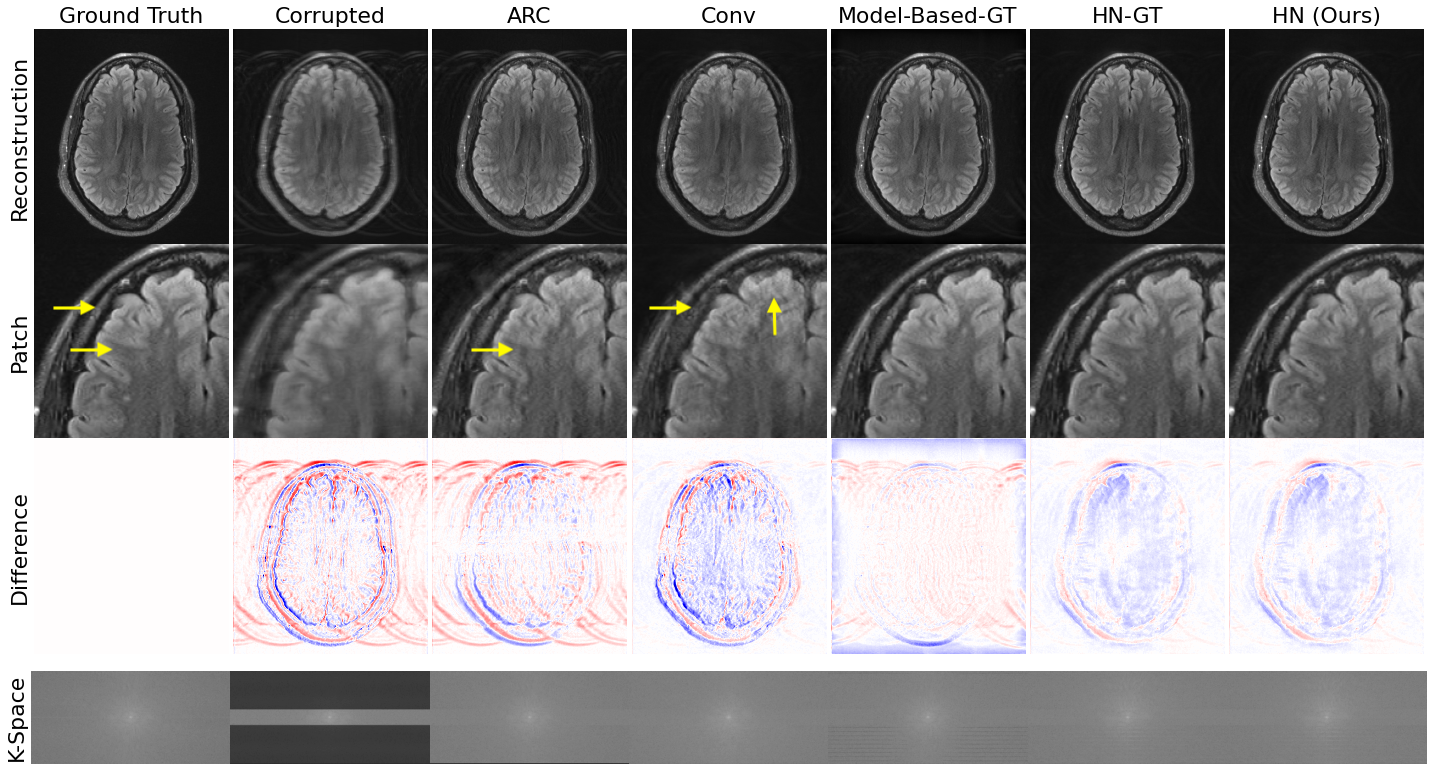
Data-consistent deep rigid motion correction for brain MRI
All dependencies required to run this code are specified in environment.yml. To create an anaconda environment with those dependencies installed, run conda env create --name <env> --file environment.yml.
You will also need to add this repo and interlacer to your python path (if you're using conda, conda-develop /path/to/neuroMoCo/).
There are three codestreams involved in our method:
- Simulation of motion-corrupted examples
- Training a model using the simulated data
- Optimizing the motion parameter estimates.
Unfortunately, we are unable to share our data and full data processing pipeline due to IRB restrictions. However, motion_sim/multicoil_motion_simulator.py contains a sample training generator which should give you an idea of how to write your own generator. Running this file will generate a folder containing .npz files used at training and test time.
The entry script to our model training is in training/train.py, which creates a model (defined in training/models.py and training/hypermodels.py) and trains the hypernetwork appropriately. training/run_experiment.py provides an example script to run training on a SLURM setup.
The test-time optimization code is found in optimization/optimize_motion_params.py, which can be run as a script to perform optimization on all the examples in a directory. Alternatively, optimize_example() can be called to run optimization on a single example.
If you use the ideas or implementation in this repository, please cite our paper:
@misc{singh2023data,
title={Data Consistent Deep Rigid MRI Motion Correction},
author={Nalini M. Singh and Neel Dey and Malte Hoffmann and Bruce Fischl and Elfar Adalsteinsson and Robert Frost and Adrian V. Dalca and Polina Golland},
year={2023},
eprint={2301.10365},
archivePrefix={arXiv},
primaryClass={eess.IV}
}