Project assignment for Bioinformatics course at University of Zagreb, Faculty of Electrical Engineering and Computing (UNIZG FER), academic year 2016/2017.
The goal was to implement fast rank/select operations on huge strings (DNA). We accomplished that by using wavelet trees with custom bitset implementation.
git clone https://github.com/filiphrenic/bioinf
cd bioinf
vagrant upUse this to clone repository and to setup environment. You don't have to run vagrant up this if you have g++-5, wget and gunzip.
bash scripts/compile.sh./bioinf [options] input_file
-o output_file [default=stdout]
-c commands_file [default=stdin]
-w word_size [default=1024]
-s [show statistics]
-v [use bool vector instead of bitmask]
-t [use red-black tree instead of lookup list]
Input file must be in FASTA format
Commands look like cmd character index; where cmd is r for rank and s for select. Example would be s A 42 which would return index of 42nd A.
./bioinf -c example/cmds_1000_1000.txt -s example/example_1000.faOr remove the -c parameter to enter commands directly via keyboard.
To run tests run
./bioinf-testchmod +x scripts/*
scripts/dl_bacteria.sh
scripts/gen_all.shThis will download DNA FASTA file of four bacterias into bacterias folder, generate synthetic files of varying lengths (100-1M) into synth folder and generate commands of varying number of commands.
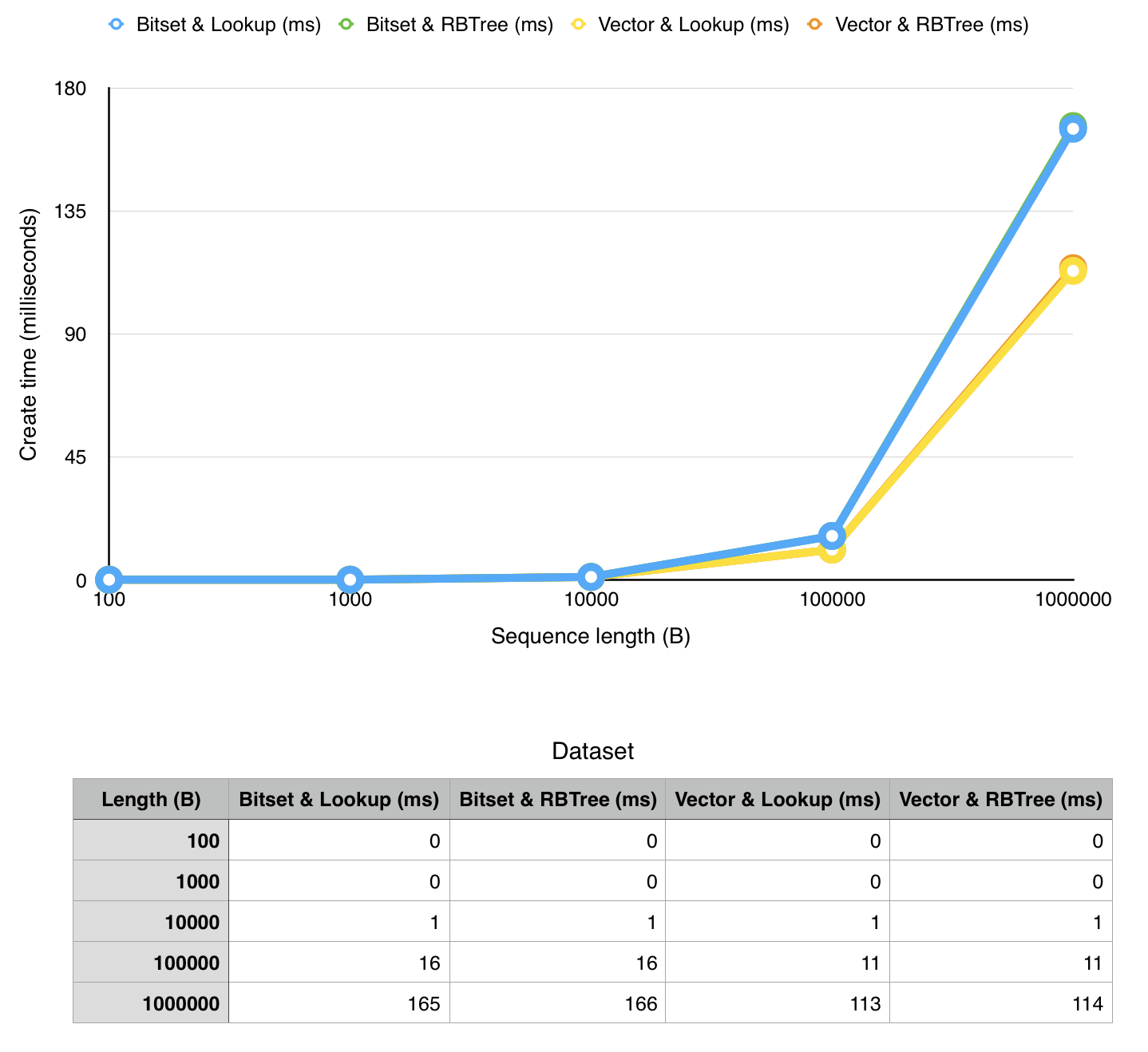
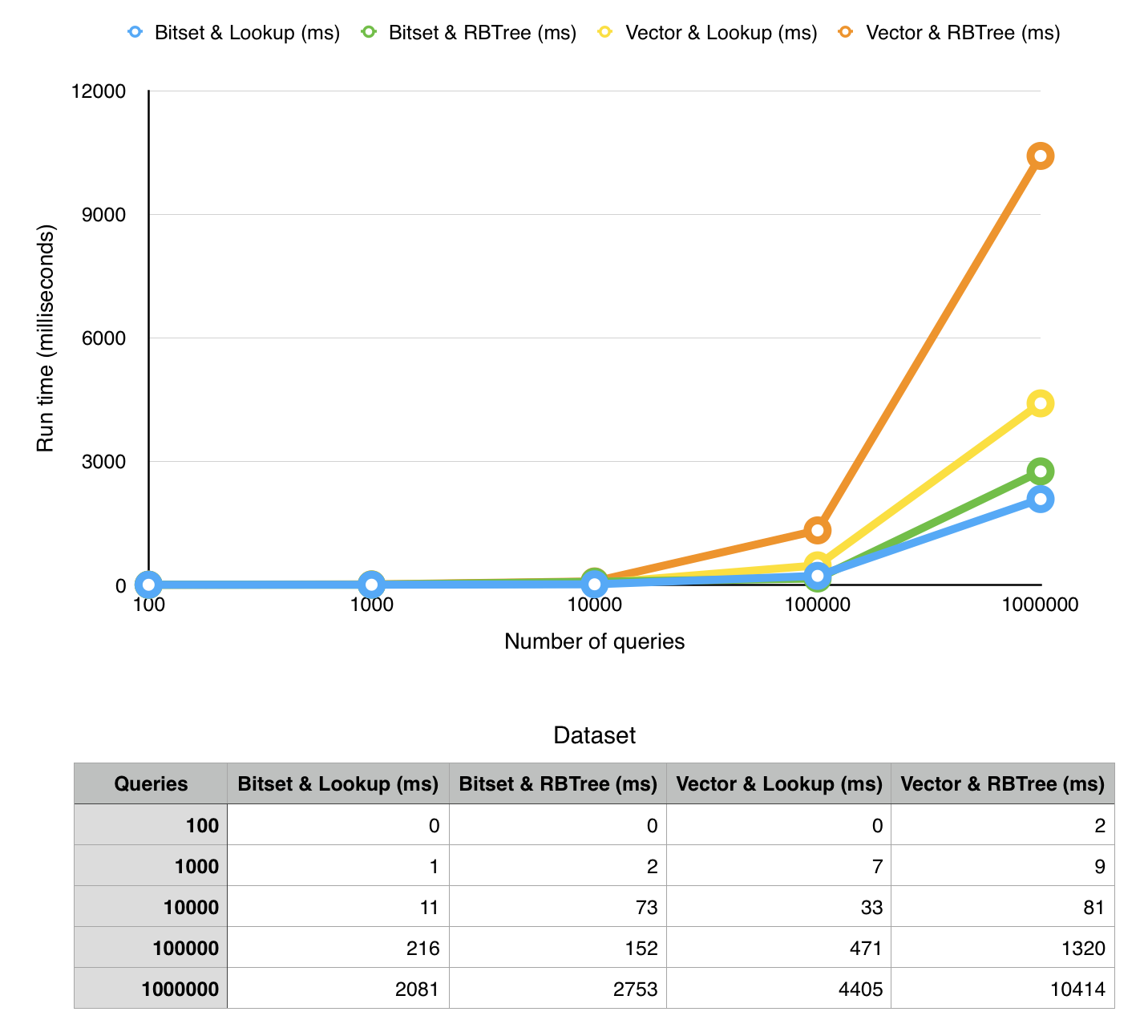
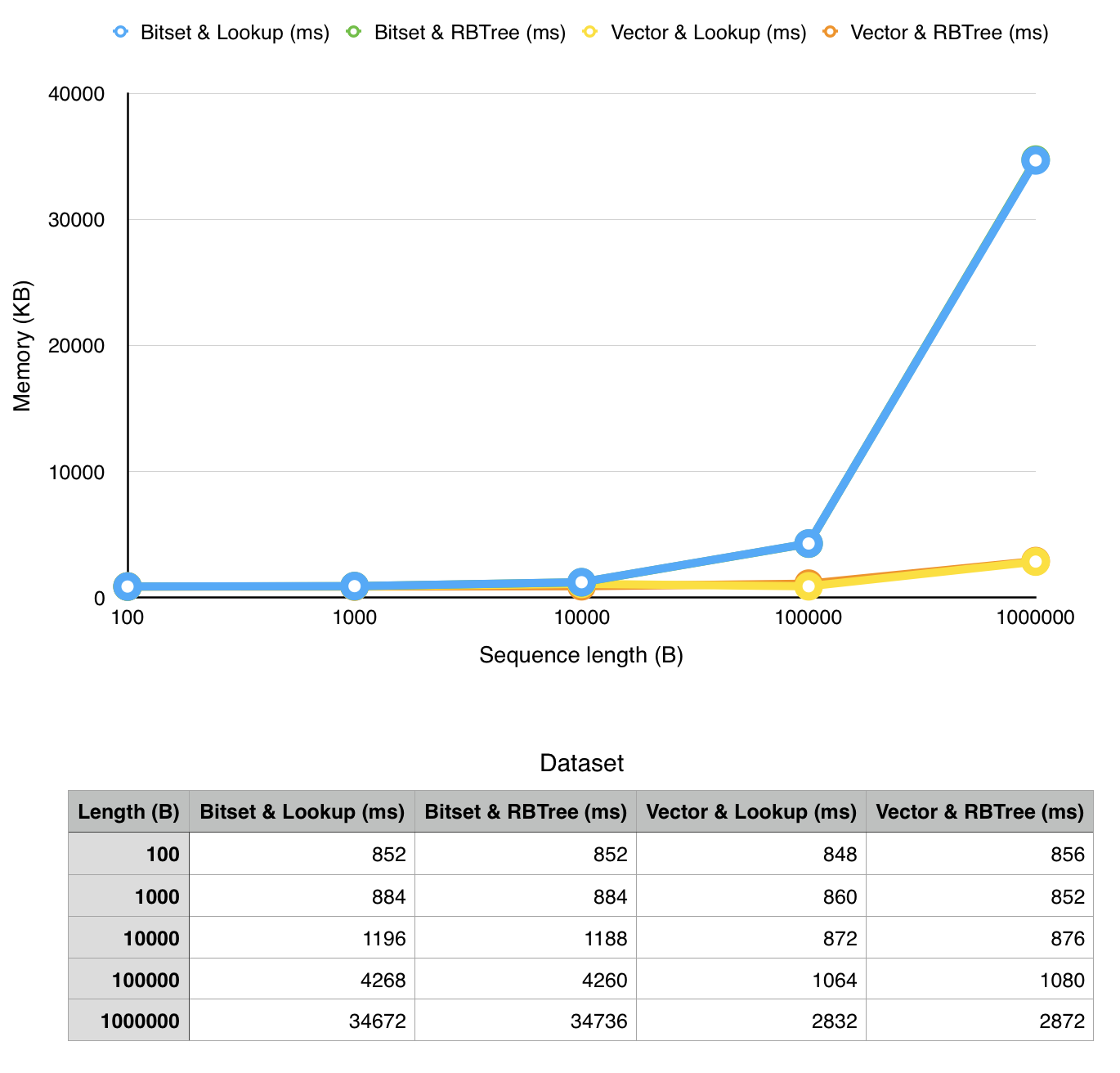
We ran thorough measurements of our implementation on a MacBook Pro 2015 (2,7 GHz Intel Core i5 with 8GB ram) in a virtual machine (Ubuntu 14.04, 4GB RAM, 2 virtual CPU's).
In the next sections we present:
- creation time (time to create data structure)
- running time (time to run a given number of queries on the structure)
- memory consumption (memory used by the process)
note that x-axis has exponential values
Definition:
* N - input file length
* W - wavelet size (1024)
* A - alphabet size (4)
* B - bitset unit size (64)
Also, let C = N/W (number of wavelets)
| Implementation | Create time | Query time | Memory |
|---|---|---|---|
| Bitset & Lookup | O(C*log(A)*(W+A)) | O(W/B*log(C)*log(A)) | O(N*log(A)) |
| Bitset & RBTree | O(C*(log(A)*(W+A)+log(C))) | O(W/B*log(C)*log(A)) | O(N*log(A)) |
| Vector & Lookup | O(C*log(A)*(W+A)) | O(W*log(C)*log(A)) | O(N*log(A)) |
| Vector & RBTree | O(C*(log(A)*(W+A)+log(C))) | O(W*log(C)*log(A)) | O(N*log(A)) |
| Species | Total length | Create time (milliseconds) | Memory (KB) |
|---|---|---|---|
| Anaplasma_phagocytophilum | 1.47M | 241 | 50620 |
| Escherichia_coli | 4.64M | 769 | 157780 |
| Mycobacterium_tuberculosis | 4.41M | 734 | 149984 |
| Salmonella_enterica | 4.95M | 814 | 168260 |
Run time depends on the number of queries and is similar to example results.
MIT License
Copyright (c) 2016 Filip Hrenić, Zvonimir Medić, Matija Oršolić