q2-mOTUs
This is a QIIME 2 wrapper for mOTU-tool. The tool will help you to assign taxonomy to your metagenomic samples. For details on QIIME 2, see https://qiime2.org.
Principal concept
mOTU is an attempt to build a taxonomy utilizing genomic information about organisms formalized with the help of differences in 40 universal gene markers (40 MGs) sequences. The basic unit of taxonomical profile is an
mOTU, and thus is used in the output. It is different from classical taxa and may encompass one, few or no species. A detailed map of relationship between mOTUs and standard taxonomical units is located in data/motus_taxonomy_map.tsv
Requirements
- QIIME 2 >= 2022.8 (https://qiime2.org/)
- Git
Known issues
QIIME2 makes the copy of data to a temporary directory. By default, it's located in the /tmp folder, which may not have enough space to store the data. Please, change the TMPDIR variable to the folder with enough space during data import.
export TMPDIR=/path/to/tmpdir
Installation
1. Install QIIME 2
Follow the instructions on https://docs.qiime2.org/2022.8/install/native/ to install QIIME 2. You will need to install the latest version of QIIME 2 (2022.8 or later).
2. Activate QIIME 2
Activate the QIIME 2 environment by running the following command:
conda activate qiime2-2022.8
3. Install mOTU-tool
Make sure to start by installing mamba in your QIIME2 environment. This will help to solve dependency conflicts faster:
conda activate qiime2-2022.8
conda install mamba -c conda-forge
Next, install q2-mOTUs
git clone https://github.com/motu-tool/q2-mOTUs
cd q2-mOTUs
make install
Test the installation
qiime dev refresh-cache
qiime motus --help
Usage
The plugin executes one function - assigns taxonomy to metagenomic reads. Therefore, there is a single workflow.
1. Import your data to QIIME 2
Import your metagenomic sequencing data in .fastq format (don't forget to preprocess your data) to QIIME2 as a SampleData semantic type using manifest file. See examples in q2_motus/tests/data.
2. Run mOTU-tool
Whether you have a single sample or multiple samples, you can run mOTU-tool using the following command:
qiime motus profile \
--i-samples q2_motus/tests/data/paired-end.qza \
--o-taxonomy paired-end-taxonomy.qza \
--o-table paired-end-classified.qza \
--p-threads 4 \
--p-jobs 2
Optimal combination threads and jobs
q2-mOTUs runs multiple instances of motu profile command from original software, which aligns reads to the reference using bwa mem. Alignment step execution time scales effectively (linearly) for up to 8 threads per job. The amount of jobs you can deploy is amount of CPUs available divided by number of threads used for a single job.
Optionaly, you can import precomputed, merged mOTU profiles
Attention: precomupted mOTU table should be generated from full taxonomy -q flag and counts -c flag profiles.
qiime motus import-table \
--i-motus-table $TMPDIR/merged.motus \
--o-table artifacts/motu-table.qza \
--o-taxonomy artifacts/motu-taxonomy.qza
Output
table-FeatureTable[Frequency]- A table of the counts of gene markers in samples.taxonomy-FeatureData[Taxonomy]- A full taxonomy for each of the gene marker.
3. Process the results
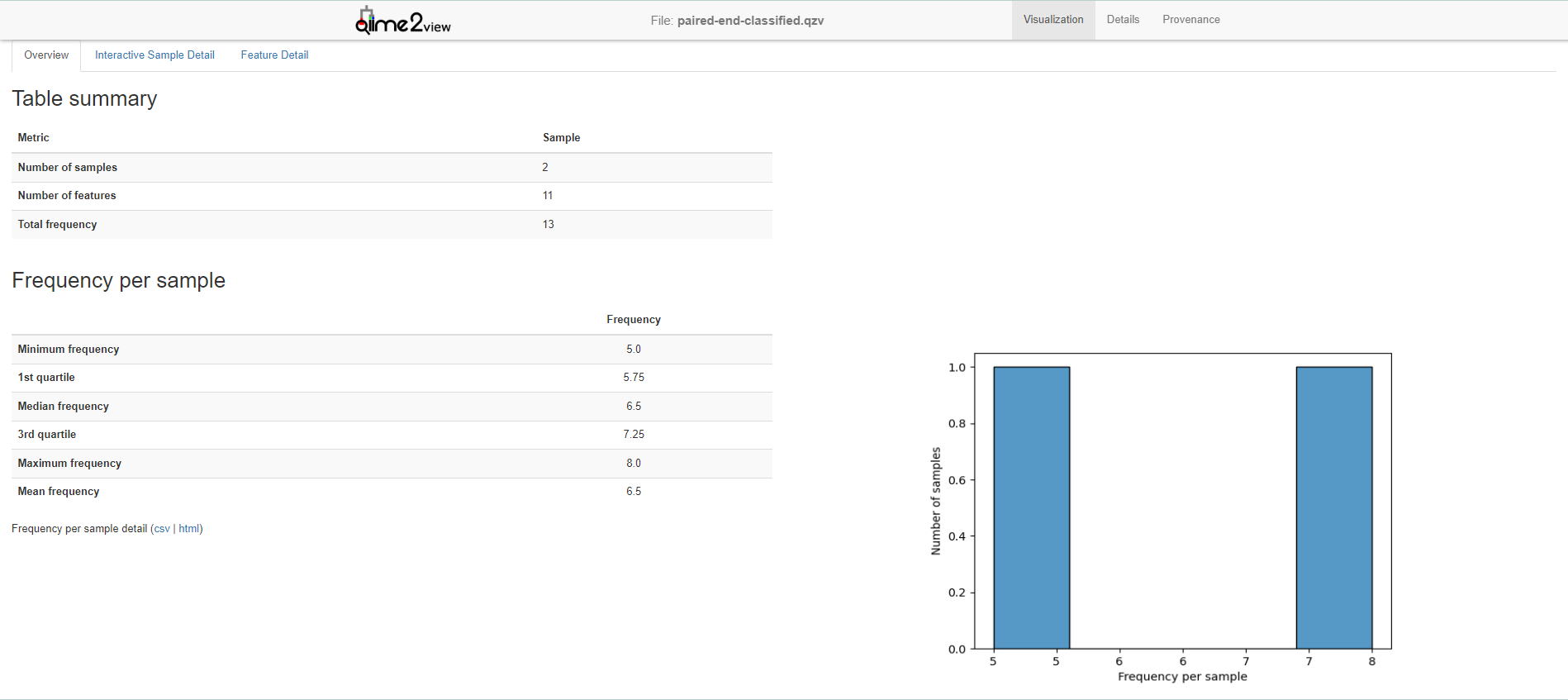
Because table is a FeatureTable[Frequency] artifact, QIIME2 offers a lot of possibilities to analyze it. For example, use feature-table summarize:
qiime feature-table summarize \
--i-table paired-end-classified.qza \
--o-visualization paired-end-classified.qzv
To get summary of your feature table.
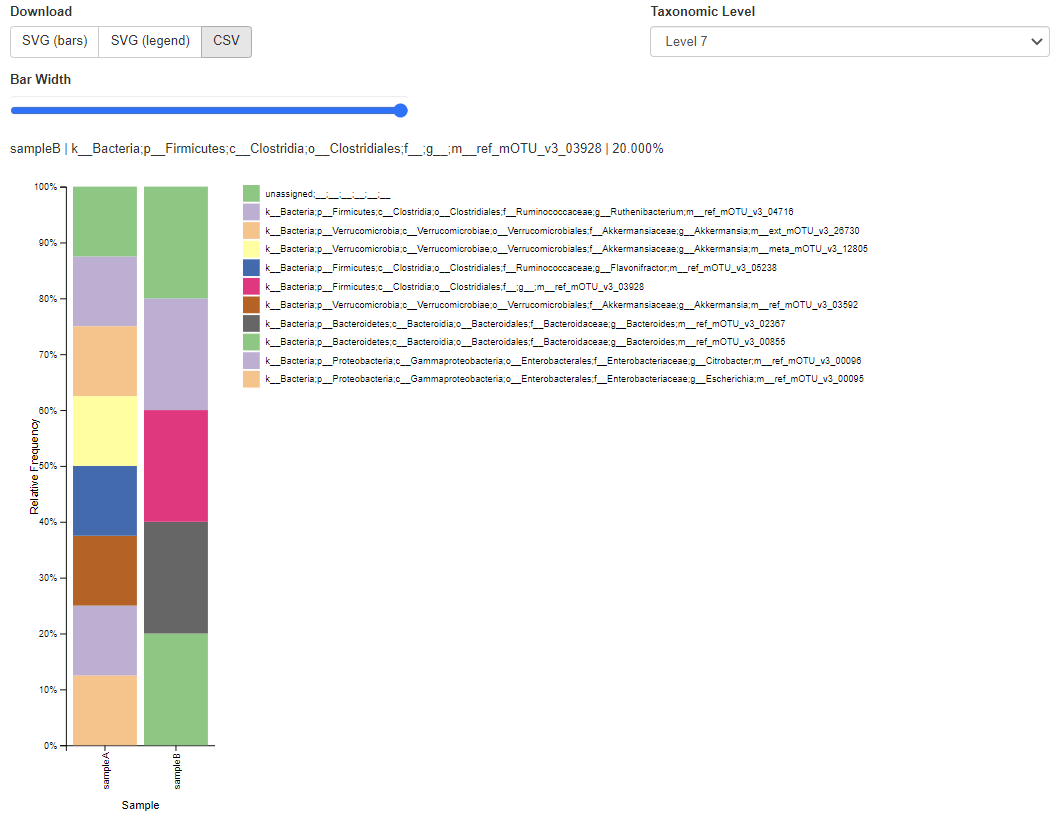
Or create all-time favourite taxa barplot:
qiime taxa barplot \
--i-table paired-end-classified.qza \
--i-taxonomy paired-end-taxonomy.qza \
--o-visualization paired-end-taxa-barplot.qzv
Or analyze the samples using Metadata you have on hand!
Parameters
Due to a QIIME2 naming convention, parameter names in plugin and standalone version are different. The table summarizes differences.
| Q2-mOTUs parameter | mOTU parameter | Description |
|---|---|---|
--p-min-alen |
-l |
Minimum length of the alignment. |
--p-marker-gene-cutoff |
-g |
Minimum number of marker genes to be considered a species. Ranges from 1 to 10. A higher value increases precision (and lowers recall). |
--p-mode |
-y |
The mode to use for abundance estimation. base.coverage measures the average base coverage of the gene. insert.raw_counts measures the number of reads that map to the gene. insert.scaled_counts measures the number of reads that map to the gene, scaled by the length of the gene. |
--p-reference-genomes/ --p-no-reference-genomes |
-e |
Only use species with reference genomes (ref-mOTUs). |
--p-threads |
-t |
Number of threads to use. |
--p-jobs |
-j |
Number of jobs to run in parallel. |
Citation
If you use this tool, please cite the following paper:
@article{Ruscheweyh2022,
doi = {10.1186/s40168-022-01410-z},
url = {https://doi.org/10.1186/s40168-022-01410-z},
year = {2022},
month = dec,
publisher = {Springer Science and Business Media {LLC}},
volume = {10},
number = {1},
author = {Hans-Joachim Ruscheweyh and Alessio Milanese and Lucas Paoli and
Nicolai Karcher and Quentin Clayssen and Marisa Isabell Keller and
Jakob Wirbel and Peer Bork and Daniel R. Mende and Georg Zeller and
Shinichi Sunagawa},
title = {Cultivation-independent genomes greatly expand taxonomic-profiling
capabilities of {mOTUs} across various environments},
journal = {Microbiome}
}