These are some preliminary results on applying Curve-GCN (https://arxiv.org/abs/1903.06874) to the problem of cardiac MRI segmentation.
Unlike commonly used neural networks for image segmentation (FCN, U-net), Curve-GCN predicts contour points around an object's boundary similar to how humans annotate images.
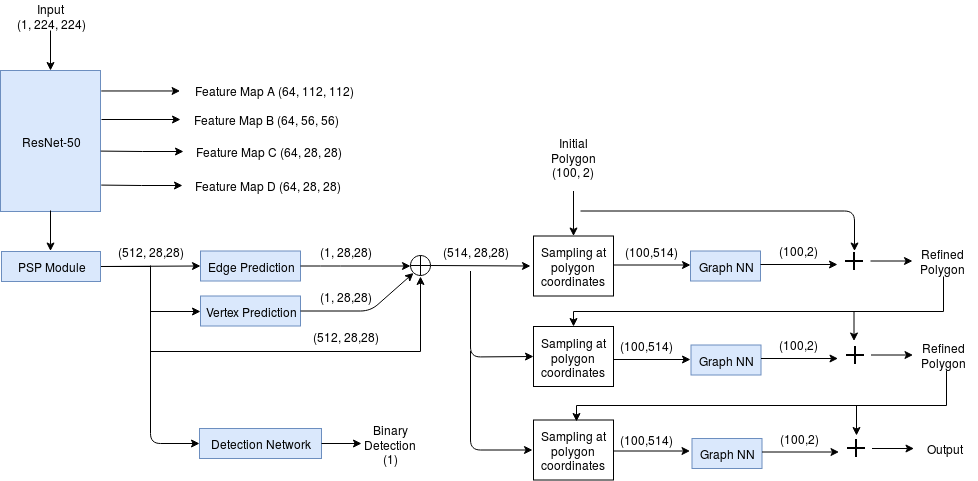
For this specific application, I extended the network to perform segmentation of upto 3 objects: left ventricle (LV), myocardium (Myo) and right ventricle (RV). Also, since some images only have 1 or 2 of these structures, I added an additional head to classify whether the image contains each structure or not.
A diagram of the network architecture is shown below.
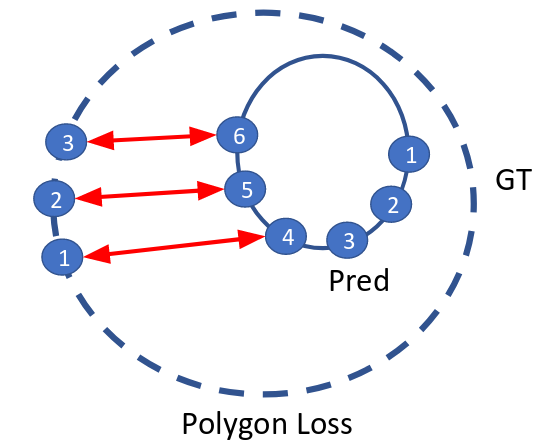
I used the polygon loss for predicting the contour loss and a binary cross-entropy loss for the additional classification branch.
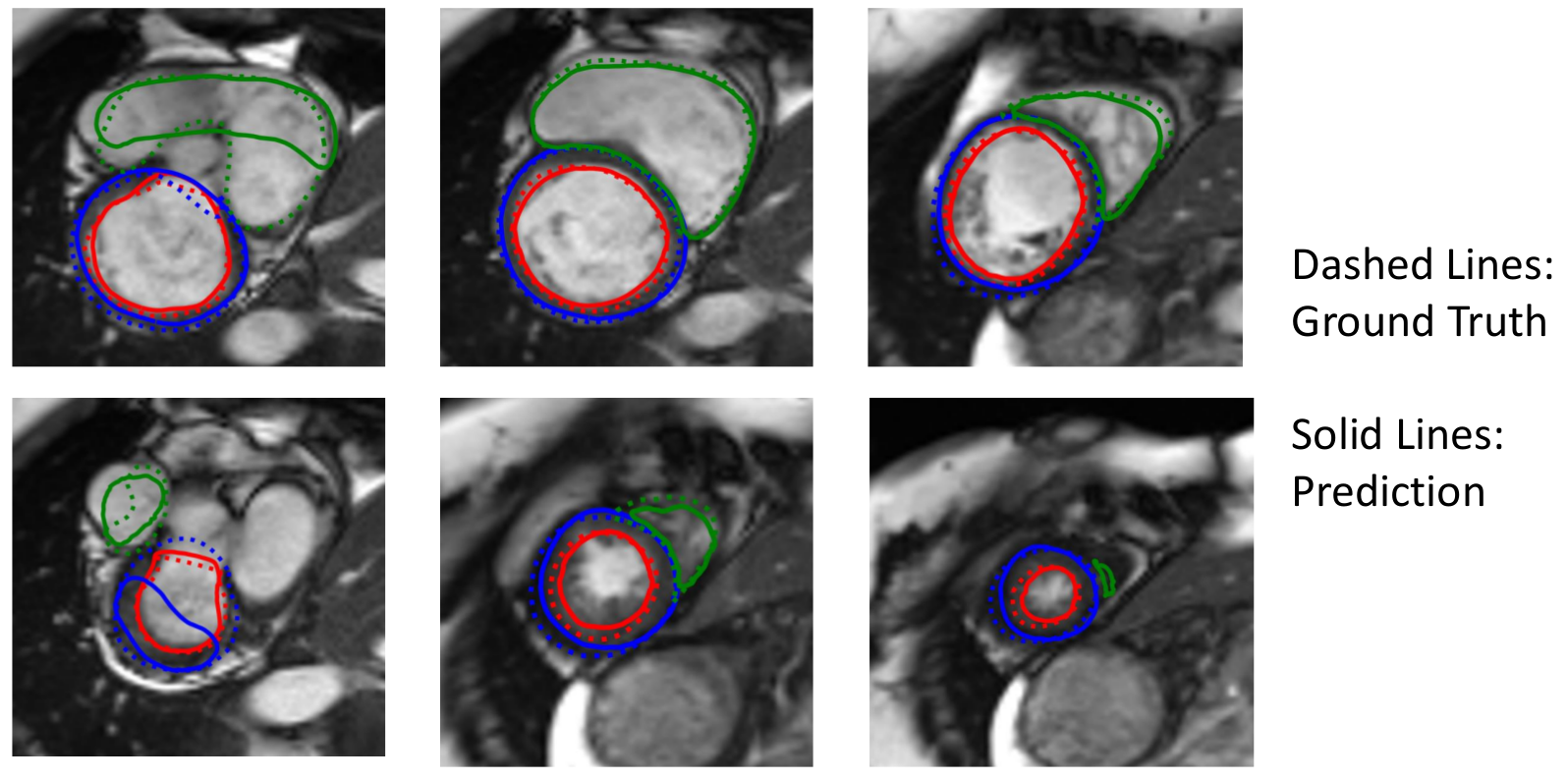
Representative results are shown below. While the LV segmentation performance is comparable to U-net performance, Myo and RV segmentation performance are worse than the U-net.
-
There is a jump from image or feature maps to coordinates. Using something like CoordConv (https://arxiv.org/abs/1807.03247) might help.
-
We are only using a small fraction of pixels in each image/feature map.
-
Add constraints on curvature of polygon
-
Add IOU/Dice loss
-
Add domain specific knowledge:
- Red contour must be inside blue contour
- Green contour shares border with blue contour on most slices
-
Sampling and GNN are not equivariant to translation
-
More data
Note that due to the original Curve-GCN code's license, a lot of the code was redacted here. Please send me an email at matthewng.ng@mail.utoronto.ca if you would like to learn more about this project.