A powerful tool for identifying non-coding RNAs in plants by analysing k-mer frequency, cds-related features, sequence length and GC content to distinguish between the growing number of non-coding RNAs and coding RNAs in plants.
- High precision (ensemble learning)
- Multiple high-performance base models
- Convenience of use
- Automated Forecasting
- Web Online
There are multiple ways to run this tool, feel free to choose one of the following method.
- Download the PINC and Add the data file to the project directory.
git clone https://github.com/midisec/PINC
cd PINC
# upload the data file (example: data.fasta)All input data must be in fasta format
- Pull and build the environment image. (Time required)
sudo docker build -t pinc_images .- Create and Enter a new container.
sudo docker run -it pinc_images bash- Execute PINC for prediction
python pinc.py -f data.fastagit clone https://github.com/midisec/PINC
cd PINC
pip3 install -r requirements.txt- Execute PINC for prediction
python pinc.py -f data.fastapython pinc.py -f <data.fasta>
After this, you will get a task page address with the uuid.
After that you can also check the history of the task by the uuid, usually it will be saved for one month.
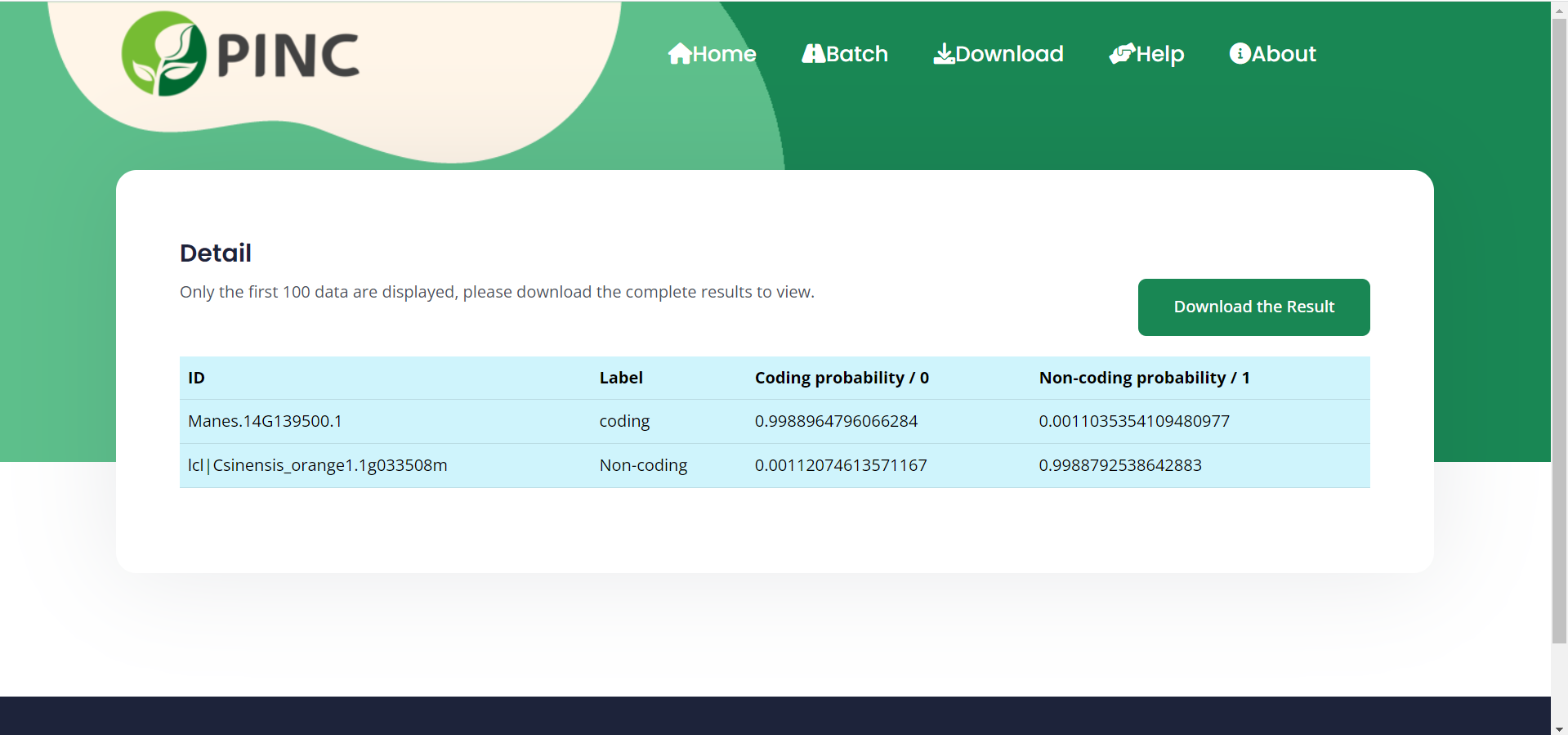
View Results and Download results
The Training set data and validation set data (7 : 3).
| Species | Coding | Non-coding | Total |
|---|---|---|---|
| Arabidopsis thaliana | 2000 | 2000 | 4000 |
| Glycine max | 2000 | 2000 | 4000 |
| Oryza sativa | 2000 | 2000 | 4000 |
| Vitis vinifera | 2000 | 2000 | 4000 |
| Total | 8000 | 8000 | 16000 |
| Species | Coding | Non-coding | Total |
|---|---|---|---|
| Cicer arietinum | 2099 | 2099 | 4198 |
| Gossypium darwinii | 5622 | 5622 | 11244 |
| Lactuca sativa | 4682 | 4682 | 9364 |
| Manihot esculenta | 2808 | 2808 | 5616 |
| Musa acuminata | 2059 | 2063 | 4122 |
| Nymphaea colorata | 1708 | 1708 | 3416 |
| Solanum tuberosum | 8282 | 8282 | 16564 |
| Sorghum bicolor | 8657 | 8657 | 17314 |
| Zea mays | 7406 | 7406 | 14812 |
| Total | 51323 | 51327 | 102650 |
In the test set, the accuracy of the PINC ranged from 92.74% to 96.42%.
@article{zhang2022pinc,
title={PINC: A Tool for Non-Coding RNA Identification in Plants Based on an Automated Machine Learning Framework},
author={Zhang, Xiaodan and Zhou, Xiaohu and Wan, Midi and Xuan, Jinxiang and Jin, Xiu and Li, Shaowen},
journal={International Journal of Molecular Sciences},
volume={23},
number={19},
pages={11825},
year={2022},
publisher={MDPI}
}