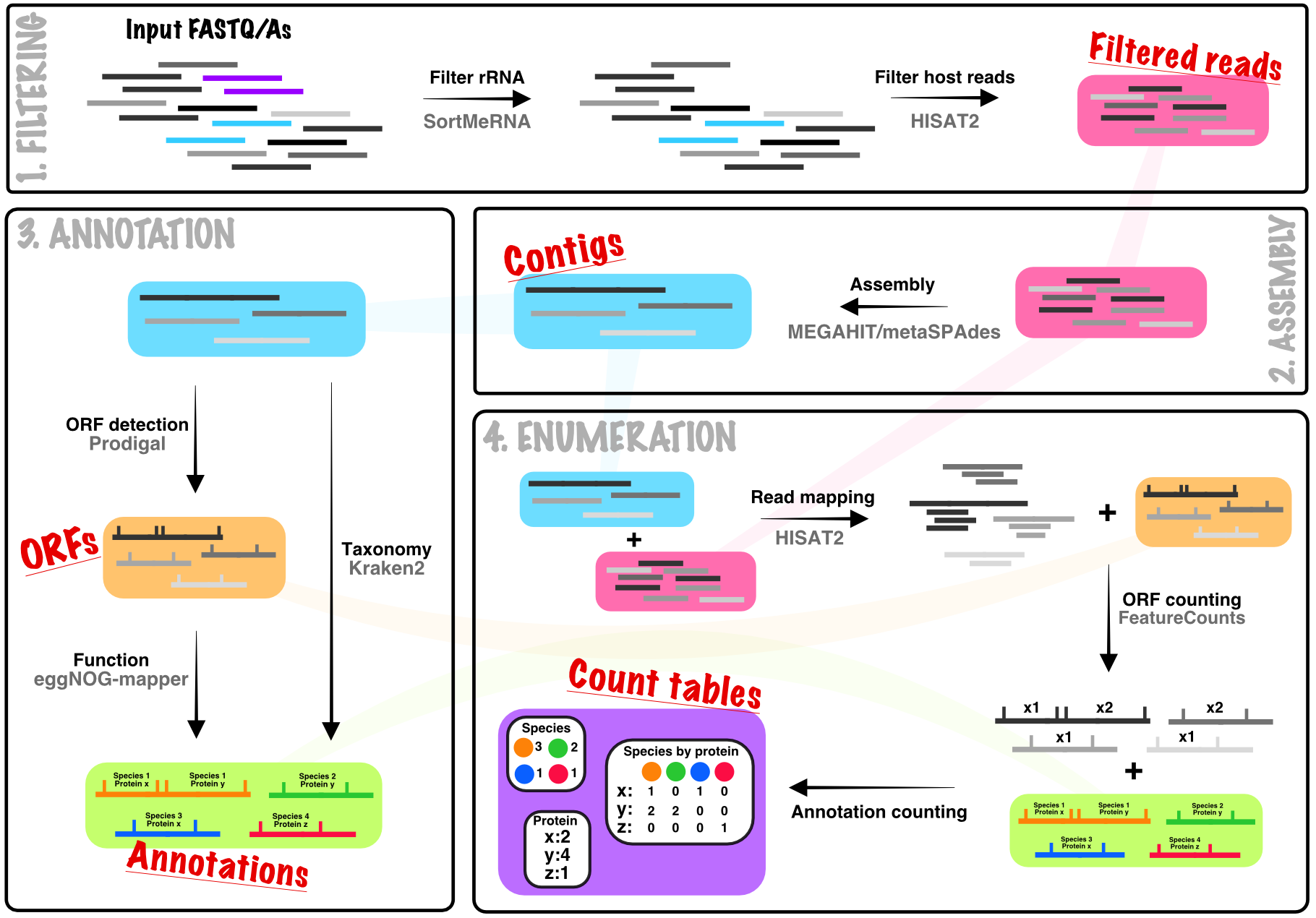
MetaSequencing takes a partial assembly approach to processing metagenomic data, assembling short-reads to contigs that are used for taxonomic annotation and de novo open-reading frame detection and functional annotation. This provides a trade-off between short-read only methods and full metagenomic genome assembly, conceptually increasing taxonomic assignment accuracy by using longer contigs whilst negating the need to deal with binning and associated issues as when trying to reconstruct full genomes. This also enables quantification of function at the per taxon level.
The pipeline can be installed into isolated Conda environments using the attached install script and full metagenomic processing can be run on FASTA/FASTQ files using a single command, generating summary reports alongside processed data tables. MetaSequencing is built using the SnakeMake workflow management system. A previous version of the pipeline based on the CGAT workflow is available here but lacks some improvements made in this version and is no longer actively developed.
Note: The complete pipeline has been tested in-house but no formal benchmarking is currently available. However, each of the individual approaches wrapped by the pipeline have been published and applied previously. Hopefully some formal testing will be done soon!
When executed in a folder containing single or paired-end FASTA/Q files for each sample in an experiment, by default, the pipeline will run all of the steps outlined below on a per sample basis. It will then collate the per sample counts into experiment-wide tables. Each of the four sub-stages (Filtering, Assembly, Annotation, and Enumeration) can also be run independently if only specific intermediates are required (see Running MetaSequencing).
MetaSequencing and its dependencies are best installed and run within a self-contained Conda environment. Following the instructions below will create the necessary environments and install all dependencies. This assumes an existing working install of Conda.
#clone this repository into the folder where you would like the install
git clone https://github.com/microbialman/MetaSequencing.git
cd MetaSequencing
#install of the main metasequencing environment may take several minutes as it solves
sh install.shNote: This will generate two conda environments (some tools require Python 2 in place of 3). If you are manually installing MetaSequencing to environments without the default names you will also need to edit the calls that load the Python 2 environment in the config .yaml files as needed
To run the full pipeline, place all the sample files in a single folder. There should be a single file or pair of files (for paired end sequencing) per sample. Paired end files should have the format SampleName.1.fastq SampleName.2.fastq. Input files can also be gzip compressed (.gz extension).
Note: If using symbolic links to files make sure they are specified using the full paths, Snakemake doesn't detect relative links.
Parameters can be changed in the .yaml files in the config directory. There are independent configuration files for each of the main stages of the pipeline. These can be used to set parameters including the reference databases used, select features to count in the enumeration step, and the threads and memory to allocate to each stage.
Altering the config directory files will make the changes global for all MetaSequencing runs. If you wish to set parameters for an individual run you can create a copy of the file and specify it with --configfile in the call to Snakemake, this will override the default parameters.
Prior to running, ensure the MetaSequencing Conda environment is active.
conda activate metasequencingThen, from the directory containing the files to be analysed, the pipeline can be called as follows:
snakemake -s /path/to/install/MetaSequencing.smkAdditional arguments to the snakemake workflow can be found in the Snakemake documentation. Useful additions to consider are highlighted below:
#this will do a dry run of the pipeline and just show what jobs will be executed
snakemake -s /path/to/install/MetaSequencing.smk -n
#this will submit the jobs to a Slurm cluster, limiting to 40 concurrent jobs
#compute cluster use is recommended, see Snakemake docs for more info on profiles for different systems
snakemake -s /path/to/install/MetaSequencing.smk --profile slurm -j 40Each stage will output into Filtering, Assembly, Annotation, and Enumeration folders. These will contain the filtered reads, contigs, annotation files and counts tables respectively. Within each folder there will also be a report html file with plots that provide an overview of the data. These can be combined into a single report that also summarises the Snakemake jobs by running the following command:
snakemake -s /path/to/install/MetaSequencing.smk --report report.htmlThe counts tables at each taxonomic level represent the summed counts of reads mapping to open reading frames assigned to each taxon. Similarly, the predicted protein names represent direct counts of how many reads mapped to ORFs assigned to that protein name. Other functional annotations, such as KEGG Pathways, can have multiple mappings (each ORF can have multiple pathway annotations). The default in this case is to split the read counts assigned to each ORF evenly across all of its annotations. An alternative approach to handle multiple annotations is to use set enrichment with the .gmt files produced by the pipeline.
Each of the four main stages can also be run individually. This can be done by editing the global.yaml file to specify which stage to run. The pipeline is then run as before in a folder containing the FASTA/Q files for processing. E.g. the annotation step is run in a folder containing the contig files to be annotated, the enumeration step in a folder of reads to be mapped.
Some additional parameters may need to be set when running steps in isolation, for example specifying the contigs and annotations for the enumeration step, these are detailed in the config files as necessary.
As it is not clear that splitting counts across multi-mapping annotations is an appropriate method to enumerate functions, the pipeline also provides methods for set enrichment analysis. Files are generated that map predicted protein names to all of the pathways/other annotations that they are assigned to. These files can then be used with the predicted protein name counts to carry out set enrichment analyses (as commonly done for RNA-Seq datasets). It also provides these mappings between taxonomic levels so that taxonomic enrichments can be similarly tested, e.g enrichment of higher taxonomic levels can be tested from species level association results.
These .gmt files can be found in the Annotation/gmt_files.dir and Annotation/taxon_gmt_files.dir folders. Once association measures have been established with base-level annotations, such as species and predicted protein names, the effect sizes/p-values can then be used to test for enrichment of higher-level annotations using gene-set enrichment tools such as Piano and fgsea in R.