Tensorflow implementation of the DGP-VAE model. Accompanying code for our paper.
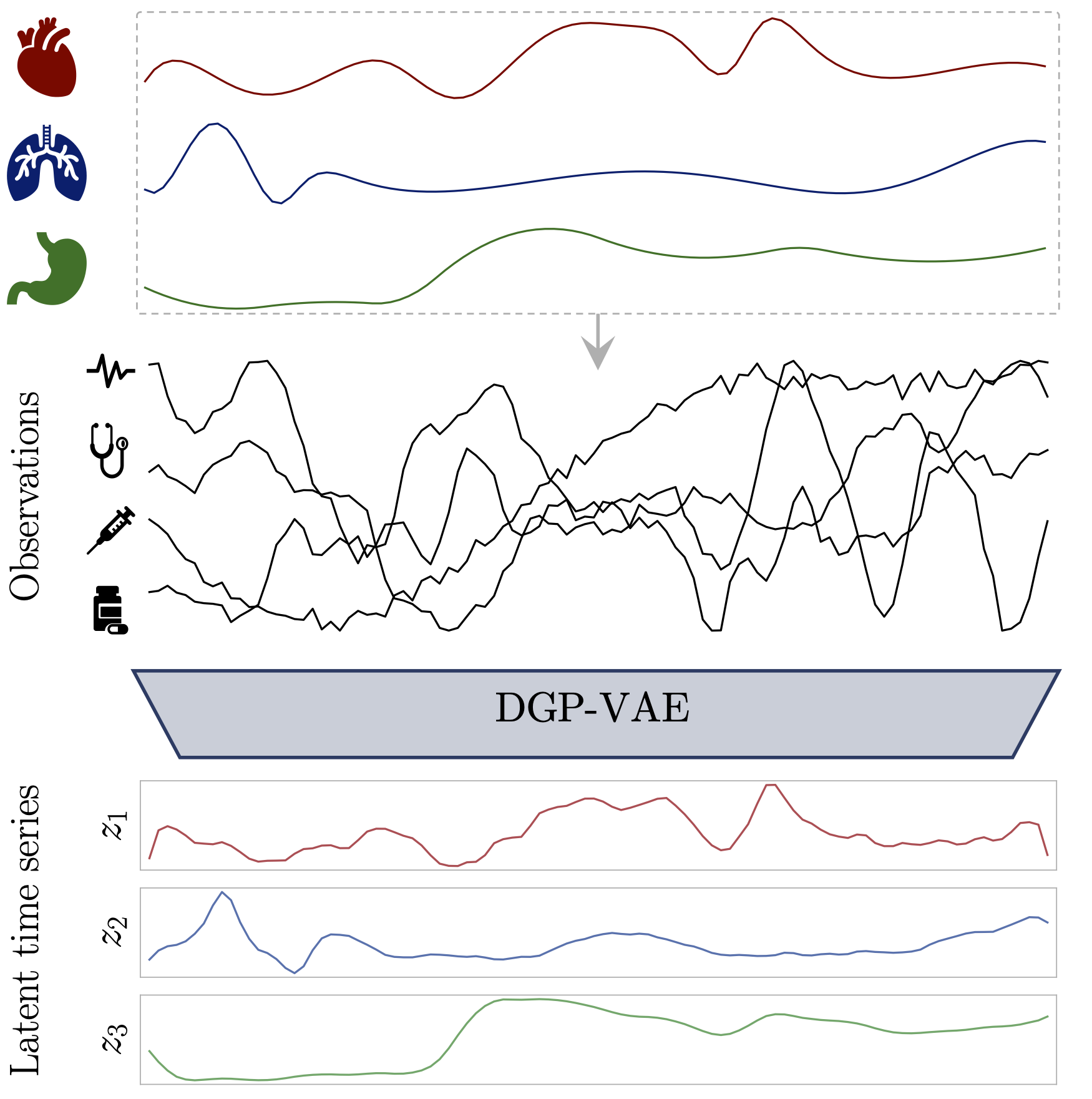
We investigate the performance of a GP-VAE type model to learning disentangled representations from time series by providing a slightly modified model: the DGP-VAE.
Our model learns disentangled representations from sequential data by modeling each independent latent channel with a Gaussian Process and employing a structured variational distribution that can capture long-term dependencies in time.
We show the efficacy of our approach in two experiments:
-
A benchmark experiment comparing against state-of-the-art disentanglement models on synthetic data.
-
An experiment on real medical time series data, where we provide a detailed comparison with the state-of-the-art model for disentanglement that can exploit the structure of sequential data.
- Python >= 3.6
- TensorFlow = 1.15
- disentanglement_lib (Available here)
- Some more packages: see
requirements.txt
- Clone or download this repo.
cdyourself to its root directory. - Grab or build a working python enviromnent. Anaconda works fine.
- Install dependencies, using
pip install -r requirements.txt - Clone or download the disentanglement_lib and add its root directory to your
PYTHONPATH. - Download data:
- Synthetic data: First, download the underlying factors of variation by running
data/load_{dsprites, smallnorb, cars3d, shapes3d}.sh. Then synthesize the actual data set from these factors by runningpython data/create_dataset.py --data_type {dsprites, smallnorb, cars3d, shapes3d}. - Real medical data: Download the HiRID data set
and save the Parquet partitons of the
mergedstage of the pre-processed data in a directory. To create the training data from the partitions, runpython create_hirid.py --hirid_merged_dir /path/to/hirid/partitions. Finally, to download the assignment matrix required for the DCI score calculation, rundata/load_hirid_assign.sh.
- Synthetic data: First, download the underlying factors of variation by running
- Run the command
python run_experiment.py --model_type dgp-vae --data_type {dsprites, smallnorb, cars3d, shapes3d, hirid} --exp_name <your_name> ...
To see all available flags run: python train.py --help
The exact hyperparameters used for the experiments are reported in our paper. To reproduce our final results run the following commands:
-
dSprites, SmallNORB, Cars3D, Shapes3D:
python run_experiment.py --model_type dgp-vae --data_type {dsprites, smallnorb, cars3d, shapes3d} --time_len 5 --testing --batch_size 32 --exp_name reproduce_{dsprites, smallnorb, cars3d, shapes3d} --seed $RANDOM --banded_covar --latent_dim 64 --encoder_sizes=32,256,256 --decoder_sizes=256,256,256 --window_size 3 --sigma 1 --length_scale 2 --beta 1.0 --data_type_dci {dsprites, smallnorb, cars3d, shapes3d} --shuffle --save_score --visualize_score -
HiRID:
python run_experiment.py --model_type dgp-vae --data_type hirid --time_len 25 --testing --batch_size 64 --exp_name reproduce_hirid --seed $RANDOM --banded_covar --latent_dim 8 --len_init scaled --kernel_scales 4 --encoder_sizes=128,128 --print_interval 1 --decoder_sizes=256,256 --window_size 12 --sigma 1.005 --length_scale 20.0 --beta 1.0 --shuffle --data_type_dci hirid --save_score --visualize_score