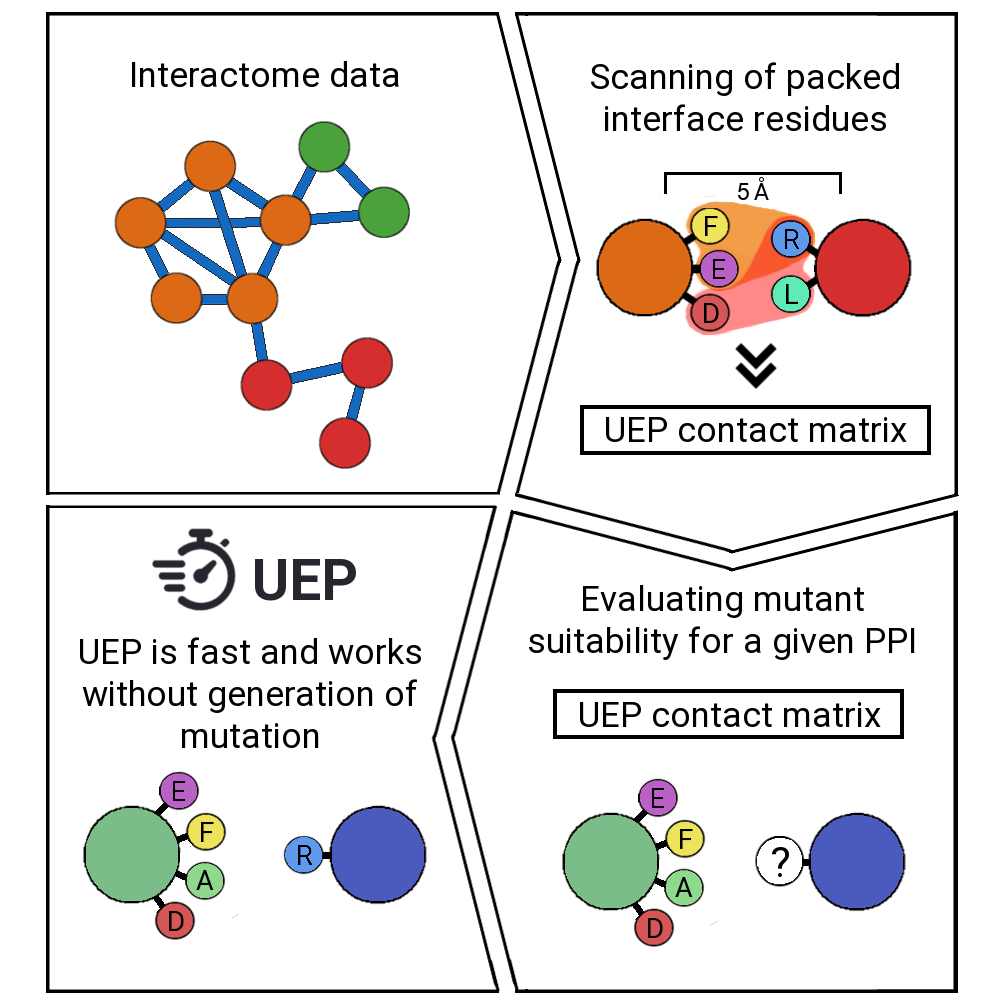
UEP is a tool for predicting the impact of mutations in a protein-protein complex. For a given PDB structure, it predicts automatically the effects of all possible mutations in the highly-packed interface positions. UEP predictions are competitive with the state-of-the art methods in the field with the advantage of being open-source code and extremelly fast in comparison, since it will take less than a second!
- argparse - user input
- prody - three-dimensional searches
- itertools - combinatorial calculations
- os - pathing files
- numpy - calculations
- compress_pickle - reading UEP contact matrix
- pandas - exporting results as csv file
- Clone UEP repository in your computer.
- Define a PDB file and an interface to work on following this scheme.
python3 UEP.py --pdb=PDB.pdb --interface=A,BC
- Results will be displayed and saved (csv file) in the same folder than your PDB path.
- Open the results file (csv file).
- First column represents analyzed positions on the highly packed interface.
- Other columns represent mutations into the different residues.
- Numerical values represent the predicted ΔΔG.
- NaN values represent mutations that could not be scored because:
- Mutation is the same residue than the wild type.
- Mutation has less than 2 predicted contacts with the other chains.
- Negative ΔΔG values are mutations predicted to improve the binding affinity of the PPI.
- Positive ΔΔG values are mutations predicted to decrease the binding affinity of the PPI.
Current state-of-the art methods for predicting the impact of mutations in a protein-protein complex rely on the description of physical energies, statistical potentials, conservation, shape complementarity, and more recently, machine learning-based approaches.
UEP moves appart from the state-of-the art and it is based on the interactions observed in the interactome data (https://interactome3d.irbbarcelona.org/). It follows a three-body contact scheme of the highly-packed positions, where one residue of one protein must be in contact with at least two residues of the other protein. We have observed that such highly-packed positions exert larger differences in the experimental ΔΔG, and therefore they i) are easier to be predicted, and ii) they are more interesting for protein-protein design campaings.
Once you run UEP, it will find the highly-packed residues of your PDB, and it will examine the contacts of your protein-protein interface. Then, it will predict a ΔΔG based on the wild type and the mutation counts observed in the interactome data, without the need of generating mutation files. This feature makes UEP really fast!