pauvre: a plotting package designed for nanopore reads
This package currently hosts one script for plotting.
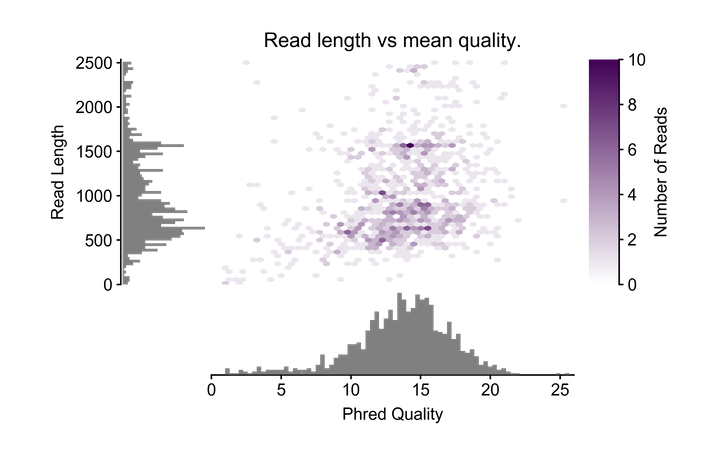
pauvre marginplot- takes a fastq file as input and outputs a marginal histogram with a heatmap.
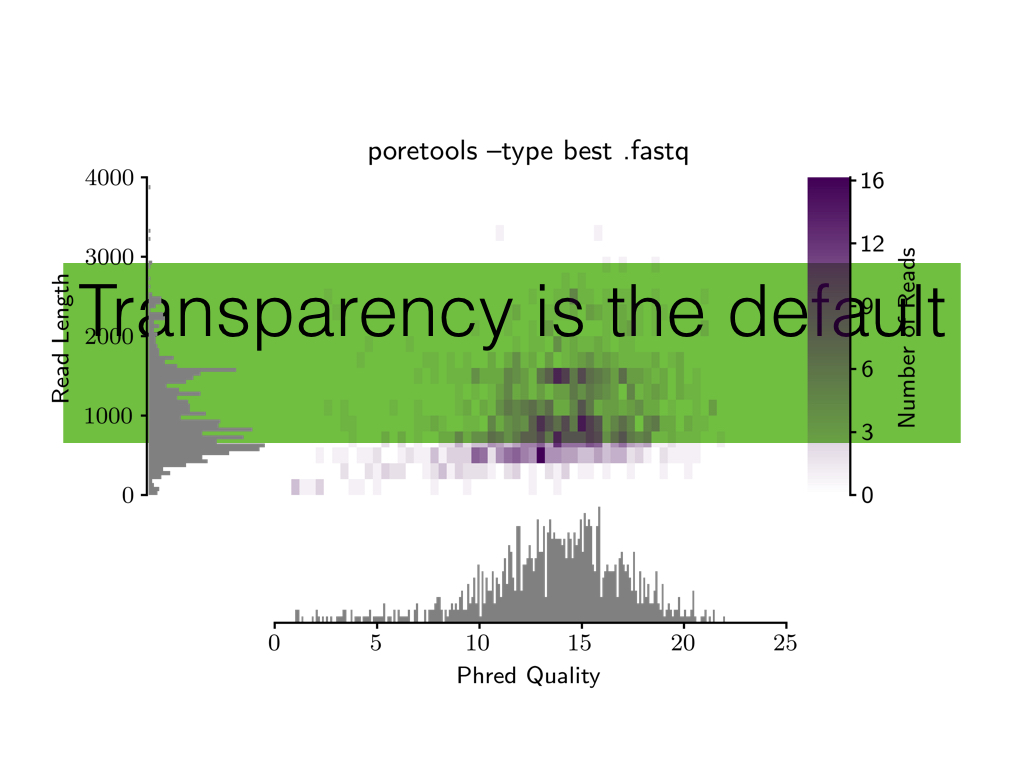
- transparency is the default
Updates:
- 20170529 - added automatic scaling to the input fastq file. It scales to show the highest read quality and the top 99th percentile of reads by length.
Requirements
- You must have the following installed on your system to install this software:
- python 3.x
- matplotlib
- biopython
- pandas
- pillow
Installation
- Instructions to install on your mac or linux system. Not sure on
Windows! Make sure python 3 is the active environment before
installing.
git clone https://github.com/conchoecia/pauvre.gitcd ./pauvrepip3 install .
- Or, install with pip
pip3 install pauvre
Usage
stats- generate basic statistics about the fastq file. For example, if I
want to know the number of bases and reads with AT LEAST a PHRED
score of 5 and AT LEAST a read length of 500, run the program as below
and look at the cells highlighted with
<braces>.pauvre marginplot --fastq miniDSMN15.fastq
- generate basic statistics about the fastq file. For example, if I
want to know the number of bases and reads with AT LEAST a PHRED
score of 5 and AT LEAST a read length of 500, run the program as below
and look at the cells highlighted with
numReads: 1000
numBasepairs: 1029114
meanLen: 1029.114
medianLen: 875.5
minLen: 11
maxLen: 5337
N50: 1278
L50: 296
Basepairs >= bin by mean PHRED and length
minLen Q0 Q5 Q10 Q15 Q17.5 Q20 Q21.5 Q25 Q25.5 Q30
0 1029114 1010681 935366 429279 143948 25139 3668 2938 2000 0
500 984212 <968653> 904787 421307 142003 24417 3668 2938 2000 0
1000 659842 649319 616788 300948 103122 17251 2000 2000 2000 0
et cetera...
Number of reads >= bin by mean Phred+Len
minLen Q0 Q5 Q10 Q15 Q17.5 Q20 Q21.5 Q25 Q25.5 Q30
0 1000 969 865 366 118 22 3 2 1 0
500 873 <859> 789 347 113 20 3 2 1 0
1000 424 418 396 187 62 11 1 1 1 0
et cetera...
marginplot- automatically calls
pauvre statsfor each fastq file - Make the default plot showing the 99th percentile of longest reads
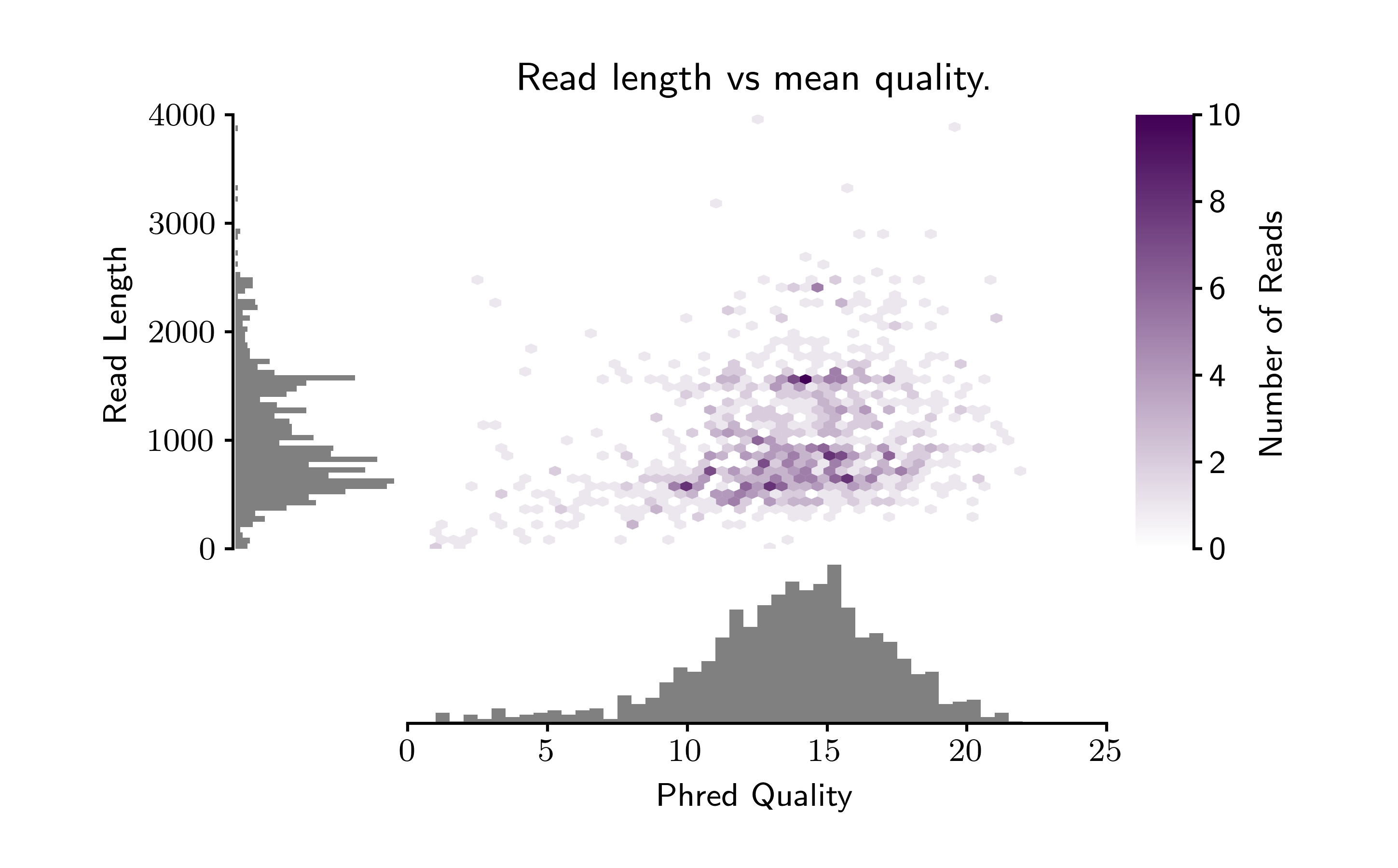
- Make a marginal histogram for ONT 2D or 1D^2 cDNA data with a lower maxlen and higher maxqual.
- Plot ONT 1D data with a large tail
pauvre marginplot --maxlen 100000 --maxqual 15 --lengthbin 500 <myfile>.fastq
- Get more resolution on lengths
pauvre marginplot --maxlen 100000 --lengthbin 5 <myfile>.fastq
- Turn off transparency if you just want a white background
- automatically calls
Contributors
@conchoecia (Darrin Schultz) @mebbert (Mark Ebbert)