Megan Barkdull
This repository hosts the workflow for a comparative genomics analysis.
If you use this code, please be sure to cite all appropriate third party tools as described in the ReadMe below, and also cite this repository using the digital object identifier (DOI) below:
If you have any questions about how to use this workflow, feel free to open a Github issue or email at mb2337 [at] cornell [dot] edu.
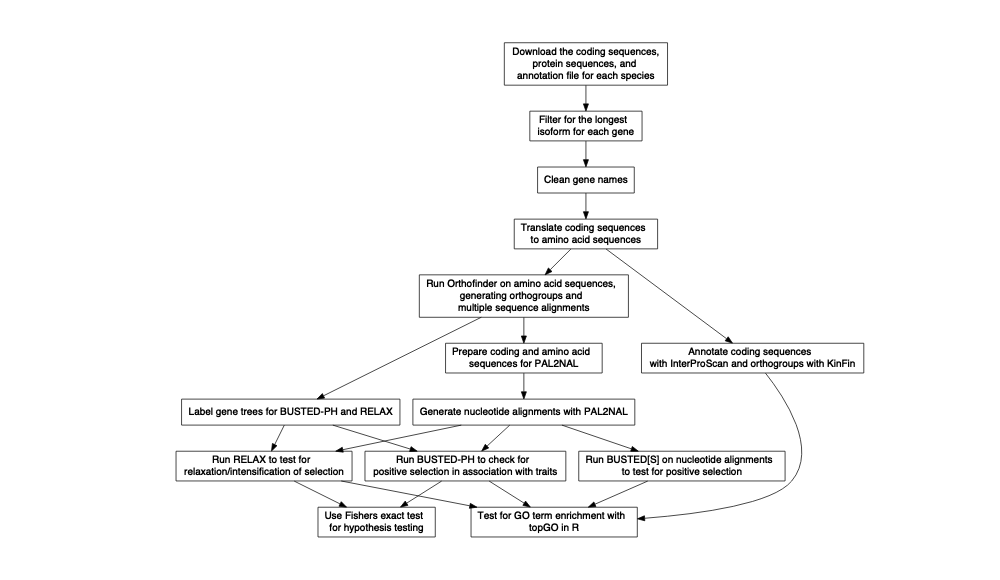
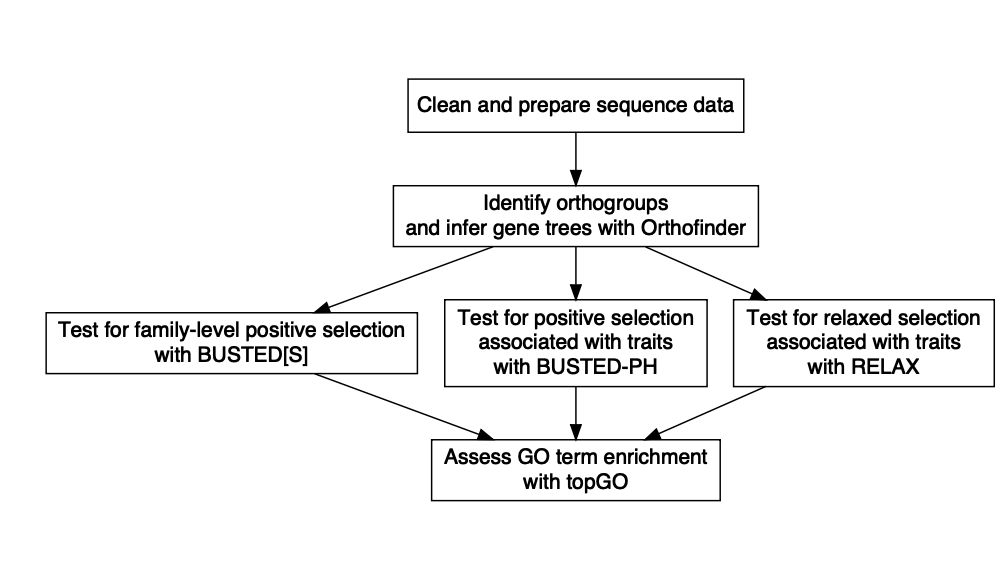
The general overview of this workflow is:
Put more simply:
To use this workflow, simply clone this repository onto your own
machine. You will run all Bash scripts in the main directory associated
with this repository (i.e., in /FormicidaeMolecularEvolution/)
For a project of this kind, you will need coding sequence and protein
data, as well as GFF-formatted annotations, from your species of
interest. The Bash script DataDownload can download these data for
you. This script requires an input file, which should be a
comma-delimited text file with:
- Coding sequence download URLs in the first column
- Protein sequence download URLs in the second column
- GFF-formatted annotation download URLs in the third column
- Species abbreviation codes in the fourth column; I suggest something like first letter of gene and first three letters of species.
- yes/no values in the fifth column; the indicates whether or not longest isoforms ought to be calculated for this species. See section 2 for details.
See ./scripts/inputurls.txt for an example.
Once you have an input URLs file, simply run the script by executing the following command in a Bash shell:
./scripts/DataDownload ./scripts/inputurls.txt
This will create a new directory, ./1_RawData, containing the
downloaded input files.
Protein and coding sequence datasets are likely to include multiple
isoforms for many genes. This project makes use of just the single
longest isoform for each gene. To filter down to just the longest
isoforms, you’ll use the script GeneRetrieval. This is an R script
which uses the R package
orthologr to filter
for longest isoforms based on a protein sequence file and a
GFF-formatted annotation file. The script then matches protein names to
gene names in order to subset the coding sequence file to just the
longest isoforms.
Crucially, in order for this script to work properly, the script expects transcript gene names to look like this:
>lcl|NW_012130065.1_cds_XP_012054525.1_1 [gene=LOC105617575] [db_xref=GeneID:105617575] [protein=proton-coupled amino acid transporter 4-like] [frame=2] [partial=5'] [protein_id=XP_012054525.1] [location=join(<125..532,740..1100,1848..2438)] [gbkey=CDS]
And processes them to look like:
XP_012054525.1
Where XP_012054525.1 is the name of a protein in the proteins file.
To run this script, simply use the command:
./scripts/GeneRetrieval /scripts/inputurls.txt
This will create two files for each species, one with the longest
protein isoforms and one with the longest coding sequence isoforms, in
the directory 2_LongestIsoforms.
If you use this step, please cite orthologr as follows:
Drost et al. 2015. Evidence for Active Maintenance of Phylotranscriptomic Hourglass Patterns in Animal and Plant Embryogenesis. Mol. Biol. Evol. 32 (5): 1221-1231. doi:10.1093/molbev/msv012
The raw data is likely to have several features that will make future
steps difficult or annoying. To solve this problem, you’ll want to run
the script DataCleaning. This will remove any special characters from
gene names (for example, _ or (), which Orthofinder will change,
causing errors) and add your four-letter taxon abbreviation to the
beginning of each gene name. To run this script, simply use the command:
./scripts/DataCleaning ./scripts/inputurls.txt
This will create a new directory, ./3_CleanedData, that contains the
cleaned transcript files.
For many steps of this workflow, you’ll actually need amino acid sequences rather than protein sequences. Therefore, we’ll need to translate the data we downloaded. This will be done in one of two ways:
- If the data you downloaded are raw transcript data, meaning for example that they don’t start with a start codon, then we’ll use Transdecoder to process and translate them into meaningful amino acid sequences.
- If they have already been processed and begin with start codons, we can take a simpler route and just translate them with the use of a codon table.
The script ./scripts/DataTranslating takes care of this process for
us. It will attempt to run
Transdecoder on
each cleaned transcripts file; if the data is unprocessed, this will
give us a translated output file. If the data is already processed,
Transdecoder will fail to run and the script will instead translate the
file with my Python script,
./scripts/TranscriptFilesTranslateScript.py. Note that the Python
script will fail in it’s entirety if it told to run on any species
that does not have data present in the directory ./3_CleanedData.
This step uses the path to Transdecoder on Cornell’s BioHPC. I plan
to update the script in future so that it is more portabe. In the
meantime, you’d need to edit lines 26 and 27 of
./scripts/DataTranslating so that they point to Transdecoder on your
machine.
To run this step, simply use the command:
./scripts/DataTranslating ./scripts/inputurls.txt
This will create a new directory, ./TranslatedData/OutputFiles/, that
contains the translated transcript files.
If you use Transdecoder, please cite it as: Haas, B., and A. Papanicolaou. “TransDecoder.” (2017).
Next, we will use Orthofinder to identify
groups of orthologous genes in our amino acid sequences, and to produce
multiple sequence alignments with MAFFT. To run Orthofinder
automatically, you can use the Bash script DataOrthofinder. This
script takes a version number as a command line option to run either
version 2.4.1 or version 2.3.8. The script is currently written to work
on Cornell’s BioHPC, and so may require some changes to run on other
machines (editing at least lines 14-16 and 42-46).
To run this step, use the command: " ./scripts/DataOrthofinder [version number, either 'current' or '2.3.8'] [full path to a species tree where tip names correspond to file names, for example 'acep_transcripts']
This will infer orthogroups and multiple-sequence alignments of amino
acid sequences; MAFFT is used to generate the multiple sequence
alignments. Outputs will be found in a new subdirectory of
/FormicidaeMolecularEvolution/, ./OrthoFinder/fasta/OrthoFinder.
If you use Orthofinder, please cite it:
- OrthoFinder’s orthogroup and ortholog inference are described here:
- Emms, D.M., Kelly, S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16, 157 (2015)
- Emms, D.M., Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20, 238 (2019)
- If you use the OrthoFinder species tree then also cite:
- Emms D.M. & Kelly S. STRIDE: Species Tree Root Inference from Gene Duplication Events (2017), Mol Biol Evol 34(12): 3267-3278
- Emms D.M. & Kelly S. STAG: Species Tree Inference from All Genes (2018), bioRxiv https://doi.org/10.1101/267914
***Please also cite MAFFT: ***
- K. Katoh, K. Misawa, K. Kuma, and T. Miyata. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14): 3059-3066.
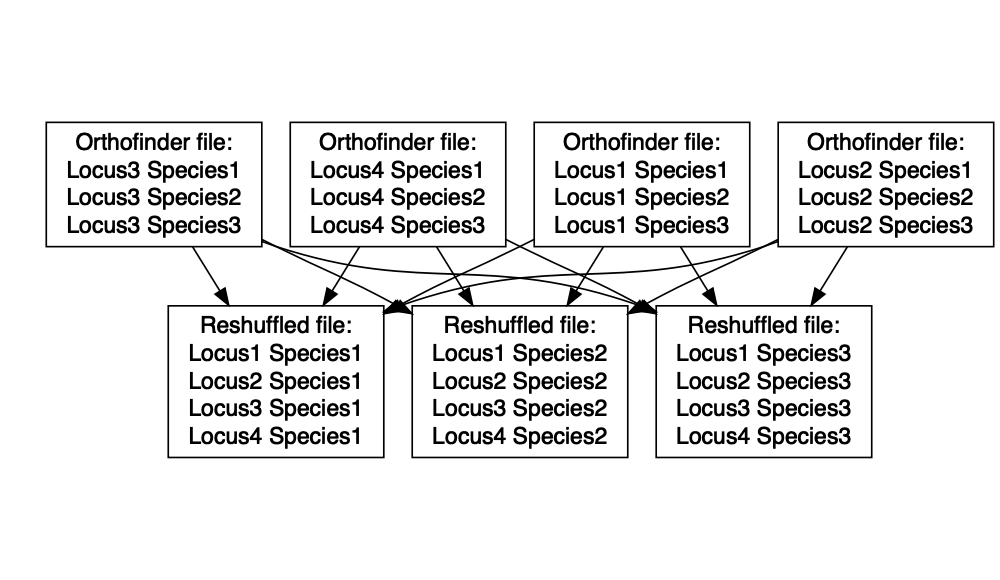
Orthofinder produces a single file for every individual orthogroup, containing the alignments for the sequences in that orthogroup. However, in order to create codon-aware alignments with PAL2NAL, we need files that contain all of the alignments for each individual species.
The R script DataMSA.R will recombine the Orthofinder outputs so that
they can be input to PAL2NAL. To run the script, you’ll need to get the
full path to the MSA files created by Orthofinder in the previous step
(it will be something like
/workdir/mb2337/FormicidaeMolecularEvolution/OrthoFinder/fasta/OrthoFinder/Results_*/MultipleSequenceAlignments).
Then use the command:
./scripts/DataMSA.R ./scripts/inputurls /FullPathToMSAFiles
The recombined output files can be found in the directory
6_1_SpeciesMSA.
The nucleotide sequence file that is run through PAL2NAL can have only
the genes that are also present in the protein alignment file. Since our
protein alignment files contain only a subset of genes, we need to
filter through the nucleotide sequence files so that they too contain
only this subset. To do this, use the R script FilteringCDSbyMSA.R.
This script checks whether genes in the coding sequence file are present
in the multiple sequence alignment file, then pulls the corresponding
nucleotide sequences from the coding sequence file into a new, filtered
output.
To run this step, just use the command: ./scripts/FilteringCDSbyMSA.R ./scripts/inputurls
This script will output filtered coding sequence files to the
subdirectory 6_2_FilteredCDS.
PAL2NAL will generate codon-aware nucleotide alignments, based on an input of an amino acid multiple sequence alignment and a nucleotide sequence. These inputs have been generated by the previous step.
Now, to run PAL2NAL, use the Bash script ./scripts/DataRunPAL2NAL by
executing the command:
./scripts/DataRunPAL2NAL ./scripts/inputurls
This will produce a new directory, 7_PAL2NALOutput, containing an
aligned nucleotide sequence file for each species.
BUSTED[S] assesses whether there is evidence for positive selection on a gene at any site along any branch in a phylogeny.
To run BUSTED[S], we will need files that contain orthologous
nucleotide sequences from each species. Therefore, we must recombine our
codon-aware alignments in a step that is the inverse of step 5.1. To do
this, use the R script ./scripts/DataSubsetCDS.R. Run with the
command:
Rscript ./scripts/DataSubsetCDS.R ./scripts/inputurls_partial [path to multiple sequence alignments from Orthofinder]
This will produce a new directory,
8_1_CDSOrthogroups/MultipleSequenceAlignments/Output that will contain
files of orthologous nucleotide sequences for input to BUSTED[S].
BUSTED[S] will not run on sequences which contain stop codons, even if
these are reasonable, terminal stop codons. Hyphy includes a utility
which will mask these these terminal stop codons in the orthogroups
(there should be few-to-no other stop codons, because our alignments are
codon-aware). To execute this step, use the script
/scripts/DataRemoveStopCodons by simply runnng the command
./scripts/DataRemoveStopCodons (no need for any additional arguments.
This will produce a new directory, 8_2_RemovedStops that will contain
files of orthologous nucleotide sequences with stop codons masked, ready
for input to BUSTED[S].
Natively, HyPhy analyses will attempt to use as many threads as
possible; however, only some steps can be run in a mult-threaded manner.
This method of running BUSTED[S] will run on a single orthogroup at a
time. To run BUSTED[S], use the Bash script
./scripts/DataRunningBusted. This script will run BUSTED[S] to test
for positive selection on the sequences. To run this step, use the
command ./scripts/DataRunningBusted [absolute path to the gene tree files from Orthofinder, something like /workdir/mb2337/FormicidaeMolecularEvolution/5_Orthofinder/fasta/Orthofinder/Results*/Resolved_Gene_Trees].
To speed up analysis, different orthogroups can be run simultaneously.
To run BUSTED[S] in this manner, use the command
./scripts/BUSTEDchunks [absolute path to the gene tree files from Orthofinder, something like /workdir/mb2337/FormicidaeMolecularEvolution/5_Orthofinder/fasta/Orthofinder/Results*/Resolved_Gene_Trees] [the number of chunks you would like to divide inputs into; likely a number near the number of cores you are working with]. I would
recommend this approach.
BUSTED[S] outputs results in the form of a JSON file. To parse these
results, you can use the script ParsingBustedResults.R. You’ll need to
have already annotated orthogroups, as described in step 11, below. This
script: + Reads in BUSTED[S] results; + Converts them from JSON files
to a spreadsheet; + Uses
topGO
to assess GO term enrichment of genes under positive selection.
To run this script, use the command:
Rscript ./scripts/ParsingBustedResults.R [full path to BUSTED[S] results]
Results will be output in the directory ./Results/.
If you use this script, please cite topGO:
- Alexa A, Rahnenfuhrer J (2021). topGO: Enrichment Analysis for Gene Ontology. R package version 2.44.0.
BUSTED-PH determines whether there is evidence for selection occuring in association with a trait/phenotype. In this workflow, we will use it to assess evidence for selection in a set of foreground branches (species of interest) vs. background branches (all other species). Like BUSTED[S], BUSTED-PH will require some data preparation steps.
BUSTED-PH
requires two things as input: a set of aligned coding sequences, and a
phylogeny- in our case, because we want to assess selection in
foreground vs. background branches, a labelled phylogeny. We have
inferred gene trees for all of our orthogroups, and now need to label
them for input to BUSTED-PH. This can be done using a script included
with HYPHY, or using the
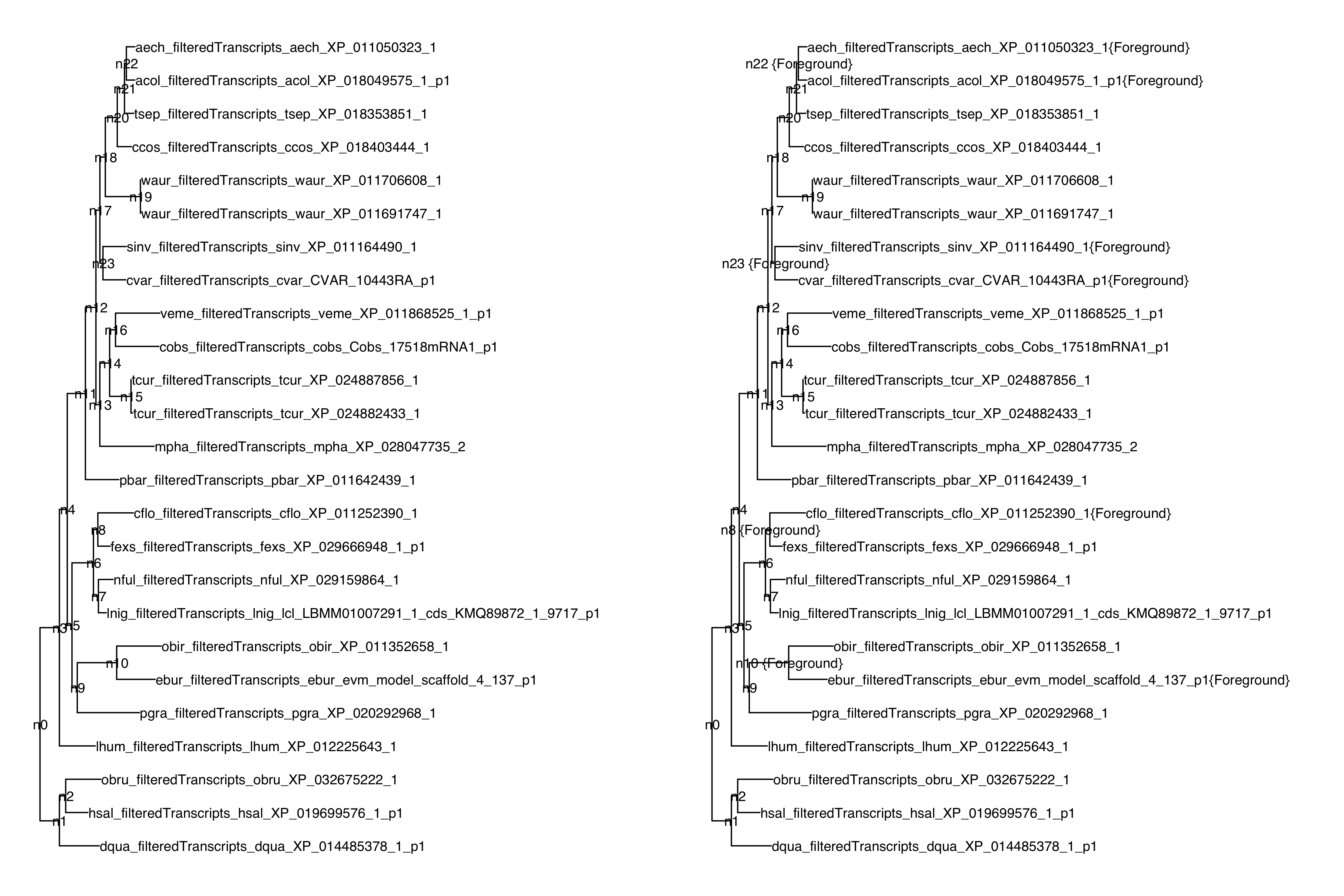
script LabellingPhylogeniesHYPHY.R. This script will label tips with
the tag {Foreground} based on their match to a set of species
abbreviations, and will label all internal nodes that are parent to
labelled tips:
LabellingPhylogeniesHYPHY.R can be run with the command:
LabellingPhylogeniesHYPHY.R [the full path to Orthofinder's gene trees] [a text file list of species abbreviations to label as foreground] [a prefix for the output files]
This will create a new directory, ./9_1_LabelledPhylogenies, with
subdirectories corresponding to each run of the script, named according
to the prefix assigned to output files of that run. In that subdirectory
will be the labelled phylogenies.
BUSTED-PH can be run with the script ./scripts/BUSTEDPHchunks. This
script subsets the orthogroups into a user-specified number of chunks,
then starts a background process for each chunk, passing them to the
script ./scripts/SingleBustedPHRun, which creates an input file and
runs BUSTED-PH. This allows you to take advantages of multiple cores on
your machine, speeding up the run. You’ll want the number of chunks you
specify on the command line to be the appropriate number of threads to
use on your machine. To run BUSTED-PH, then, use the
command:
./scripts/BUSTEDPHchunks [full path to the directory containing labelled phylogenies from step 9.1] [prefix used when labelling phylogenies] [number of threads to use/chunks to create]
Results will be output in the directory
./9_3_BustedPHResults/[labelling prefix]
BUSTED-PH is currently an unpublished method, with no paper to cite. Please check the GitHub to see if the method has been published, and cite accordingly.
I am interesting in whether there are more or fewer genes under positive
selection in association with particular traits, and in the categories
of gene that are under selection. To assess these things, you can use
the R script ParsingBustedPHResults.R. In order to run this analysis,
you’ll need to have already annotated orthogroups as described in step
11, below. This script:
- Reads in BUSTED-PH results;
- Converts them from JSON files to a spreadsheet;
- Runs a Fisher’s exact test to determine there is a signficiant difference in positive selection between the fore- and background species;
- Uses topGO to assess GO term enrichment of positively selected genes.
To run this script, use the command:
Rscript ./scripts/ParsingBustedPHResults.R [full path to BUSTED-PH results] [labelling prefix for this trait]
Results will be output in the directory ./Results/[labelling prefix].
If you use this script, please cite topGO:
- Alexa A, Rahnenfuhrer J (2021). topGO: Enrichment Analysis for Gene Ontology. R package version 2.44.0.
As with aBSREL, RELAX requires labelled phylogenies. You can generate
these with LabellingPhylogeniesHYPHY.R as described in section 9.1;
indeed, you can likely reuse the phylogenies you generated for aBSREL.
RELAX is a method from HYPHY that asks whether the strength of natural
selection has been increased or decreased along test branches. In order
to run RELAX, as with BUSTED-PH, you’ll need a labelled phylogeny and a
set of orthogroup sequences. Then, RELAX can be run with the command
./scripts/RELAXchunks [full path to labelled phylogenies] [prefix used when labelling phylogenies] [number of threads to use].
This script works in the same way as the script used to run BUSTED-PH, described above.
I am interesting in whether there are more or fewer genes under relaxed
selection in association with particular traits, and in the categories
of gene that are under relaxed selection. To assess these things, you
can use the R script ParsingBustedPHResults.R. In order to run this
analysis, you’ll need to have already annotated orthogroups as described
in step 11, below. This script:
- Reads in RELAX results;
- Converts them from JSON files to a spreadsheet;
- Runs a Fisher’s exact test, following Schneider, Adams, and Elmer (2019), to determine there is a signficiant difference in the rate of relaxed vs. intensified selection on the foreground species;
- Uses topGO to assess GO term enrichment of genes under relaxed selection.
To run this script, use the command:
Rscript ./scripts/ParsingRelaxResults.R [full path to BUSTED-PH results] [labelling prefix for this trait]
Results will be output in the directory ./Results/[labelling prefix].
If you use this script, please cite topGO:
- Alexa A, Rahnenfuhrer J (2021). topGO: Enrichment Analysis for Gene Ontology. R package version 2.44.0.
If you’re interested in drawing conclusions about the function of genes evolving under a particular selection regime, you’ll need to get functional annotations. This can be accomplished with a combination of InterProScan, which will annotate individual genes, and KinFin, which will use gene-level annotations to assign a function to whole orthogroups (which are the level of analysis for BUSTED, BUSTED-PH and RELAX).
You can run both InterProScan and KinFin with the script
RunningInterProScan, using the
command:
./scripts/RunningInterProScan [path to the input urls file] [full path to the Orthofinder fasta directory, something like inputurls_full.txt /workdir/mb2337/FormicidaeMolecularEvolution/5_OrthoFinder/fasta/]
This will output three files in the directory ./11_InterProScan:
cluster_domain_annotation.Pfam.txt,
cluster_domain_annotation.IPR.txt, and
cluster_domain_annotation.GO.txt, which can be combined with the
results of tests for selection in order to understand which genes are
under a particular selective regime.
For the step where you must convert the outputs of OrthoFinder to be inputs for RERConverge, please see the Comparative Genomics repository