Tutorial on named capture regular expressions in R and Python
In this 60 minute tutorial I will explain how to use named capture regular expressions to extract data from several different kinds structured text data.
Motivation for using named capture regular expressions, 5 minutes
Why would you want to use named capture regular expressions? They are useful when you want to extract groups of substrings from text data which has some structure, but no consistent delimiter such as tabs or commas between groups. They make it easy to convert such loosely structured text data into regular CSV/TSV data.
- The regular expression
5 foo barmatches any string that contains5 foo baras a substring. - The regular expression
foo|barmatches any string that containsfooorbar. The vertical bar indicates alternation – if any one of the options is present, then there is a match. - Square brackets are used to indicate a character class. The
regular expression
[0-9] foo barmeans match any digit, followed by a space, followed byfoo bar. - A capturing regular expression includes parentheses for extracting
data when there is a match. For example if we apply the regular
expression
([0-9]) (foo|bar)to the stringprefix 8 foo suffix, we put8in the first capture group andfooin the second. - A named capture regular expression includes group names. For
example if we apply the regular expression
(?<number>[0-9]) (?<string>foo|bar)to the stringprefix 8 foo suffix, we put8in the capture group namednumber, andfooin the capture group namedstring.
Named capture regular expressions are better than simple capturing regular expressions, since you can refer to the extracted data by name rather than by an arbitrary index. That results in code that is a bit more verbose, but much easier to understand. For example in Python,
import re
subject = 'chr10:213,054,000-213,055,000'
# Without named capture:
group_tuple = re.search("(chr.*?):(.*?)-([0-9,]*)", subject).groups()
print group_tuple[1]
# With named capture:
group_dict = re.search(r"""
(?P<chrom>chr.*?)
:
(?P<chromStart>.*?)
-
(?P<chromEnd>[0-9,]*)
""", subject, re.VERBOSE).groupdict()
print group_dict["chromStart"]Both print statements show the same thing, but the intent of the second is clearer for two reasons:
- The group names in the regular expression serve to document their
purpose. Regular expressions have a bad reputation as a write-only
language but named capture can be used to make them more readable:
“Hmmm… what was the second group
.*?supposed to match? Oh yeah, the chromStart!” - We can extract the data by group name (chromStart) rather than an arbitrary index (1), clarifying the intent of the Python code.
History, 5 minutes
| Who | When | First |
|---|---|---|
| Kleene | 1956 | Regular expression on paper |
| Thompson | 1968 | Regular expression in a program |
| Thompson | 1974 | grep |
| Wall | 1994 | Perl5 (? extensions |
| Hazel | 1997 | PCRE |
| Kuchling et al | 1997 | Named capture in Python1.5 |
| R core | 2002 | PCRE in R |
| Hazel | 2003 | Named capture in PCRE |
| Hocking | 2011 | Named capture in R |
Regular sets and regular expressions were introduced on paper by
Stephen Cole Kleene in 1956 (including the “Kleene star” * for zero
or more). Among the first uses of a regular expression in a program
was Ken Thompson (Bachelors 1965, Masters 1966, UC Berkeley) for his
version of the QED (1968) and ed (1969) text editors, developed at
Bell Labs for Unix. In ed, g/re/p means “Global Regular Expression
Print,” which gave the name to the grep program, also written by
Thompson (1974). I’m not sure about the origin of capture groups, but
Friedl claimed that “The regular expressions supported by grep and
other early tools were quite limited…grep’s capturing metacharacters
were \(...\), with unescaped parenthesies representing literal
text.” Larry Wall wrote Perl version 1 in 1987 while working at Unisys
Corporation, and it had capturing regular expressions. Perl version 5
in 1994 introduced many extensions using the (? notation. Sources:
wikipedia:Regular_expression and “A Casual Stroll Across the Regex
Landscape,” in Ch.3 of Friedl’s book Mastering Regular Expressions.
Philip Hazel started writing the Perl-Compatible Regular Expressions (PCRE) library for the exim mail program in 1997. Python used PCRE starting with version 1.5 in 1997. Source: Python-1.5/Misc/HISTORY.
From 1.5a3 to 1.5a4...
- A completely new re.py module is provided (thanks to Andrew
Kuchling, Tim Peters and Jeffrey Ollie) which uses Philip Hazel's
"pcre" re compiler and engine.
Python 1.5 introduced named capture groups and the (?P<name>subpattern)
syntax. Source: Python-1.5/Doc/libre.tex.
\item[\code{(?P<\var{name}>...)}] Similar to regular parentheses, but
the text matched by the group is accessible via the symbolic group
name \var{name}.PCRE support for named capture was introduced in 2003. Source: PCRE changelog (my copy).
Version 4.0 17-Feb-03...
36. Added support for named subpatterns. The Python syntax (?P<name>...) is
used to name a group. Names consist of alphanumerics and underscores, and must
be unique. Back references use the syntax (?P=name) and recursive calls use
(?P>name) which is a PCRE extension to the Python extension. Groups still have
numbers.
R includes PCRE starting with version 1.6.0 in 2002. Source: R-src/NEWS.1.
CHANGES IN R VERSION 1.6.0...
o grep(), (g)sub() and regexpr() have a new argument `perl'
which if TRUE uses Perl-style regexps from PCRE (if installed).
I wrote the code in https://svn.r-project.org/R/trunk/src/main/grep.c which implements named capture regular expression support for R. It was merged into base R in 2011 https://bugs.r-project.org/bugzilla3/show_bug.cgi?id=14518, and has been included with every copy of R since version 2.14.
Current usage in R and Python, 10 minutes
For a subject S (R character vector or Python pandas Series) and a
regular expression pattern P (string),
| base R function | R namedCapture package | Python pandas | returns |
|---|---|---|---|
regexpr(P, S) | str_match_named(S, P) | S.str.extract(P) | one match per subject |
gregexpr(P, S) | str_match_all_named(S, P) | S.str.findall(P) | several matches per subject |
Notes:
regexprandgregexprin R need the perl=TRUE argument for named capture.S.str.findallin Python pandas does not return capture group names.
Named capture in R
Base R supports named capture regular expressions via C code that
interfaces the Perl-Compatible Regular Expressions (PCRE) library.
The base functions regexpr and gregexpr use PCRE when given the
perl=TRUE argument. The first argument is the pattern, a single
regular expression (character vector of length 1), and the second
argument is the character vector of subjects (strings to
parse). However their output is a bunch of integers and group names,
so I wrote some functions in the namedCapture package that return
character matrices or data.frames, with column names as defined in the
named groups of the regular expression. To install the namedCapture
package, run the following commands in R:
if(!require(devtools))install.packages("devtools")
devtools::install_github("tdhock/namedCapture")Notes on related functions/packages:
regexecandregmatchesin base R implement extracting capture groups but theregexecman page indicates that perl=TRUE (and thus named capture) is not implemented.stringr::str_matchandstringi::stri_matchimplement extracting capture groups, but the stringi package does not support named capture yet as such a feature set is still considered as experimental in ICU.- https://github.com/tdhock/revector provides fast C code for a vector of named capture regular expressions (base R only provides functions for a single regular expression).
Named capture in Python
Note that in Python a P is required after the initial question mark
of each group: (?P<name>pattern).
The re module implements named capture regular expression support
via the M.groupdict() method for a match object M.
The pandas module for data analysis has some support for named
capture regular expressions. To install pandas execute one of the
following shell commands:
pip install pandas
easy_install pandasFor an instance S of the Series class, pandas provides the excellent
S.str.extract method which is the analog of str_match_named in
R. However the analog of str_match_all_named seems to be
S.str.findall, which does not support named capture.
Some examples, 30 minutes
| code | functions |
|---|---|
| chr.pos.R | str_match_named, str_match_all_named, gsub |
| differences_from_R.py | re.search, re.compile |
| chr_pos.py | str.extract, str.findall, re.subn |
| qsub-out.R | str_match_named |
| trackDb.R | str_match_all_named |
Questions from the audience, 10 minutes
How do you ever extracted data from text files? Show us how you extracted some data from a particular text file, and we will try to suggest improvements.
Coding projects: implementing functions for named capture
Project 1: I wrote the Python function str_match_named to be
analogous to the R function. To my knowledge there is no analog for
str_match_all_named in Python. Implement a function that inputs a
list of subject strings and outputs a list of matches per subject
(each list should contain zero or more dicts, one for each match).
Project 2: As of pandas 0.16.2 there is no str.extractall method
which I expect should return a Series with the same length as the
input/subject Series. Each of its elements should be a DataFrame with
a row for each match, and a column for each named group. Exercise for
the reader: fork pandas, add the str_extractall function to
https://github.com/pydata/pandas/blob/master/pandas/core/strings.py,
and submit them a Pull Request, being careful to follow
their guidelines for code contributions.
Project 3: Russ Cox’s ”Regular Expression Matching Can Be Simple And
Fast” explains that due to backreference support, several common
regular expression engines can have an exponential runtime. One way to
achieve a speedup is to drop backreference support and use the re2 C++
library, which supports named capture. Write an R package with a
function str_match_re2 that uses the RE2::PartialMatch C++ function
to obtain a match matrix like the output of str_match_named
(GSOC2016 project proposal). Add str_match_re2 to the benchmark code
below.
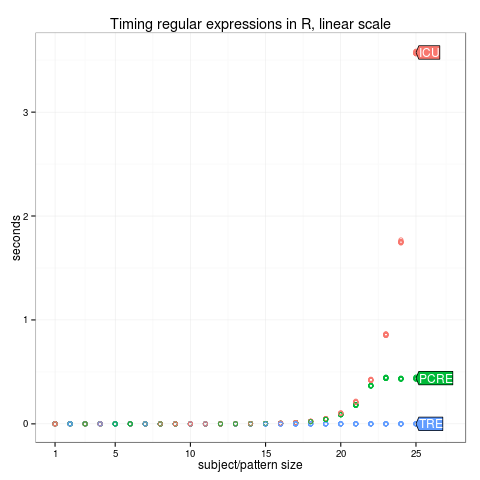
max.N <- 25
times.list <- list()
for(N in 1:max.N){
cat(sprintf("subject/pattern size %4d / %4d\n", N, max.N))
subject <- paste(rep("a", N), collapse="")
pattern <- paste(rep(c("a?", "a"), each=N), collapse="")
N.times <- microbenchmark::microbenchmark(
ICU=stringi::stri_match(subject, regex=pattern),
PCRE=regexpr(pattern, subject, perl=TRUE),
TRE=regexpr(pattern, subject, perl=FALSE),
times=10)
times.list[[N]] <- data.frame(N, N.times)
}
times <- do.call(rbind, times.list)
save(times, file="times.RData")
library(ggplot2)
library(directlabels)
linear.legend <- ggplot()+
ggtitle("Timing regular expressions in R, linear scale")+
scale_y_continuous("seconds")+
scale_x_continuous("subject/pattern size",
limits=c(1, 27),
breaks=c(1, 5, 10, 15, 20, 25))+
geom_point(aes(N, time/1e9, color=expr),
shape=1,
data=times)
(linear.dl <- direct.label(linear.legend, "last.polygons"))
png("figure-complexity-linear.png")
print(linear.dl)
dev.off()
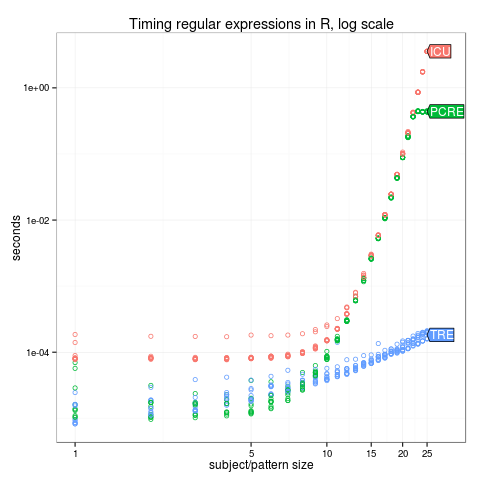
log.legend <- ggplot()+
ggtitle("Timing regular expressions in R, log scale")+
scale_y_log10("seconds")+
scale_x_log10("subject/pattern size",
limits=c(1, 30),
breaks=c(1, 5, 10, 15, 20, 25))+
geom_point(aes(N, time/1e9, color=expr),
shape=1,
data=times)
(log.dl <- direct.label(log.legend, "last.polygons"))
png("figure-complexity-log.png")
print(log.dl)
dev.off()Does your result agree with the predictions in the complexity column of the table below?
| R function | library | named capture | complexity |
|---|---|---|---|
| regexpr(perl=FALSE) | TRE | no | polynomial |
| stringi::stri_match() | ICU | no | exponential |
| regexpr(perl=TRUE) | PCRE | yes | exponential |
| str_match_re2() | re2 | yes | polynomial |
References
http://www.regular-expressions.info has some basic reference on how to write regular expressions in several languages. However it discusses neither named capture in R, nor pandas in Python.
The definitive reference on current regular expression implementations is the book “Mastering Regular Expressions,” by Jeffrey E.F. Freidl. It contains a discussion of Python and named capture but does not specifically discuss R.